Tali Mazor
Tali Mazor
I'm pretty sure we've had this issue before and that it will be fixed properly when we go live with SV, but I'm also pretty sure in the past we've...
I'm not sure if there's an example in the public portal of a sample that only has clinical data, but we have some in our internal portal. For these patients,...
I'm not sure if these are data or UI, but I noticed a few things looking at the HTAN patient timeline: 1) Labtest/IHC tooltips don't include the gene:  2)...
Labels for the rows of the heatmap are only allowed to be a single letter in a circle. This is ok when the groups are in rows, since each group...
Related to #6759, we should also add a chart for SV data, such that you can select a gene and get a chart of all the fusion partner genes for...
Similar to 'priority' for clinical attributes, I want to be able to specify in the data files for a study that certain generic assay features should be displayed in study...
The XvY charts an awesome addition to study view, but I'd love for them to support more data types. Right now they can only pull from clinical attributes, but we...
The resource data definition file has a 'description' field, but this doesn't appear anywhere in the portal. Can this be added as a column in the Resource table? 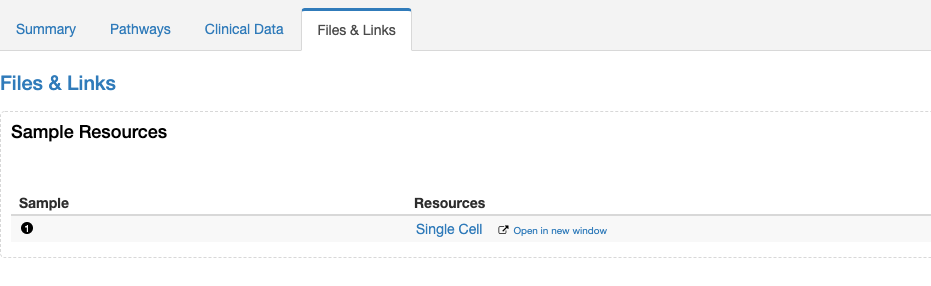
lgggbm_tcga_pub ('Merged Cohort of LGG and GBM (TCGA, Cell 2016)') includes data from both LGG & GBM cohorts, so should appear as redundant when any of those are selected. Currently...
patients 36, 47, 58 show '-' or 'antisense fusion' but should show the gene partners as they do in prod prod:  beta:  https://beta.cbioportal.org/results/download?cancer_study_list=msk_impact_2017&tab_index=tab_visualize&case_set_id=msk_impact_2017_all&Action=Submit&gene_list=TMPRSS2%250AERG%250AEGFR%250AEWSR1%250AALK%250ABRAF%250AROS1%250AEML4&show_samples=true&plots_horz_selection=%7B%22dataType%22%3A%22COPY_NUMBER_ALTERATION%22%2C%22selectedDataSourceOption%22%3A%22cna%22%2C%22mutationCountBy%22%3A%22MutationType%22%2C%22structuralVariantCountBy%22%3A%22MutatedVsWildType%22%2C%22logScale%22%3A%22false%22%7D&plots_vert_selection=%7B%22dataType%22%3A%22STRUCTURAL_VARIANT%22%2C%22selectedDataSourceOption%22%3A%22structural_variants%22%2C%22mutationCountBy%22%3A%22MutationType%22%2C%22structuralVariantCountBy%22%3A%22MutatedVsWildType%22%2C%22logScale%22%3A%22false%22%7D&plots_coloring_selection=%7B%7D&mutations_gene=EML4&comparison_subtab=alterations&comparison_selectedEnrichmentEventTypes=%5B%22structural_variant%22%5D
