Sarah Lutteropp
Sarah Lutteropp
Ok I submitted random trees and different brlen linkages experiments to haswell. The scrambled partitions stuff is more tricky, I first need to code it.
Do we also need experiments for each of the highly experimental alternative search options I implemented? Scrambling the inferred network and restarting a search from that one, extreme greedy move...
There is a problem with scaled branch lengths: ``` terminate called after throwing an instance of 'std::runtime_error' what(): I believe this function currently does not work correctly with scaled branch...
I believe my problem with scaled branch lengths was that if I already multiply the branch length by the scaler, do I also need to multiply the likelihood derivatives with...
In case someone is interested in it: This is the function where I was unsure about how to correctly handle the scaler: https://github.com/lutteropp/NetRAX/blob/master/src/likelihood/LikelihoodDerivatives.cpp#L30
Another problem with the experiments: So far, I just simulated a single network for each combination of n_taxa and n_reticulations. We need yet another experiment (linked brlens, raxml-ng best tree)...
I will be posting the merged result CLV files here: https://github.com/lutteropp/NetRAX/issues/81
That's the tree inferred by raxml-ng, which I will use as start network for NetRAX: 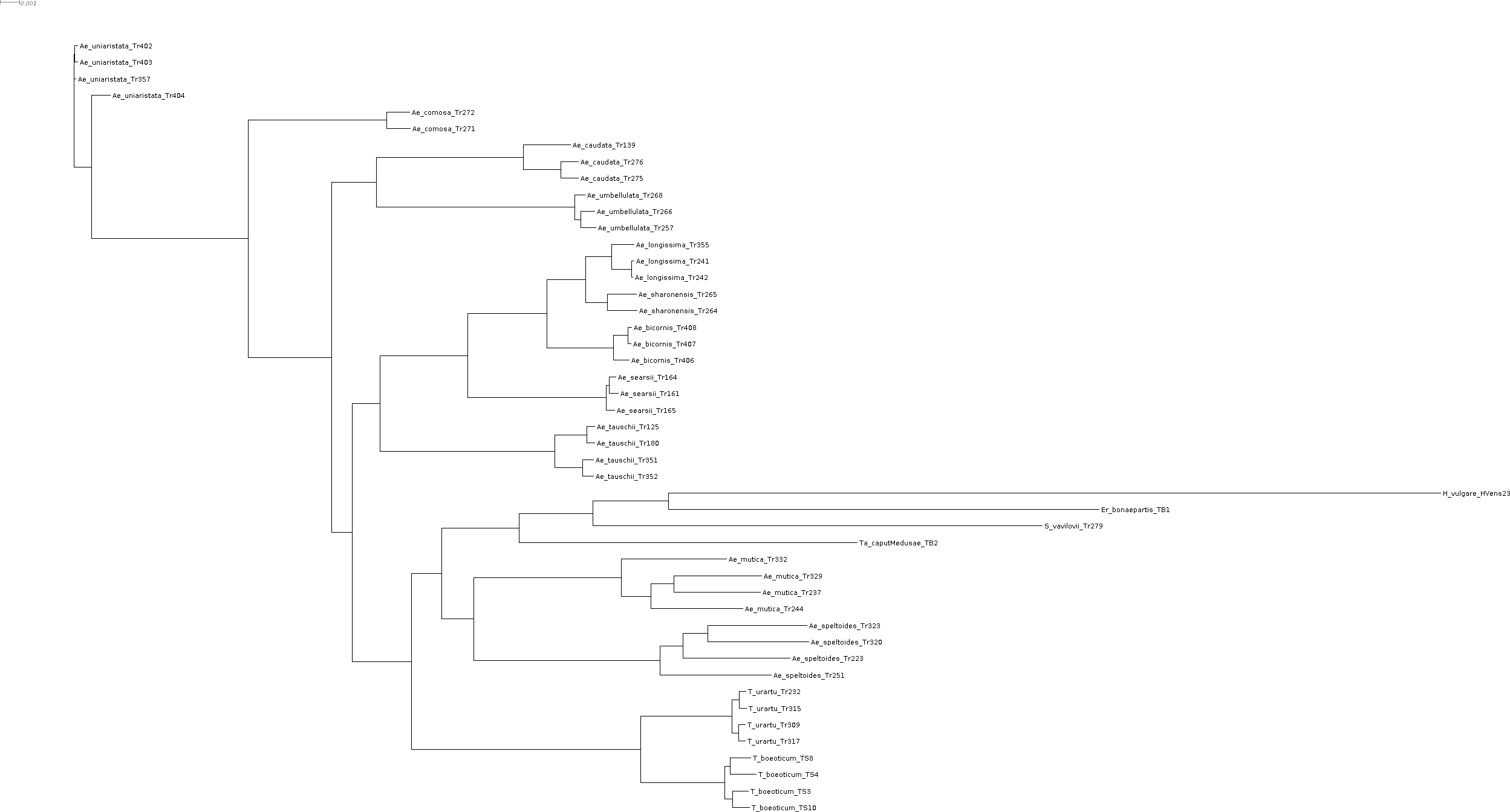 [merged_genes.raxml.bestTree.txt](https://github.com/lutteropp/NetRAX/files/6428815/merged_genes.raxml.bestTree.txt) raxml-ng vanilla (with 10 random and 10 parsimony starting trees, site repeats...
This is the best 3-reticulation network we found so far, with running NetRAX under the Likelihood.BEST linked brlens model on the complete empirical dataset, using the RAxML-NG bestTree as single...
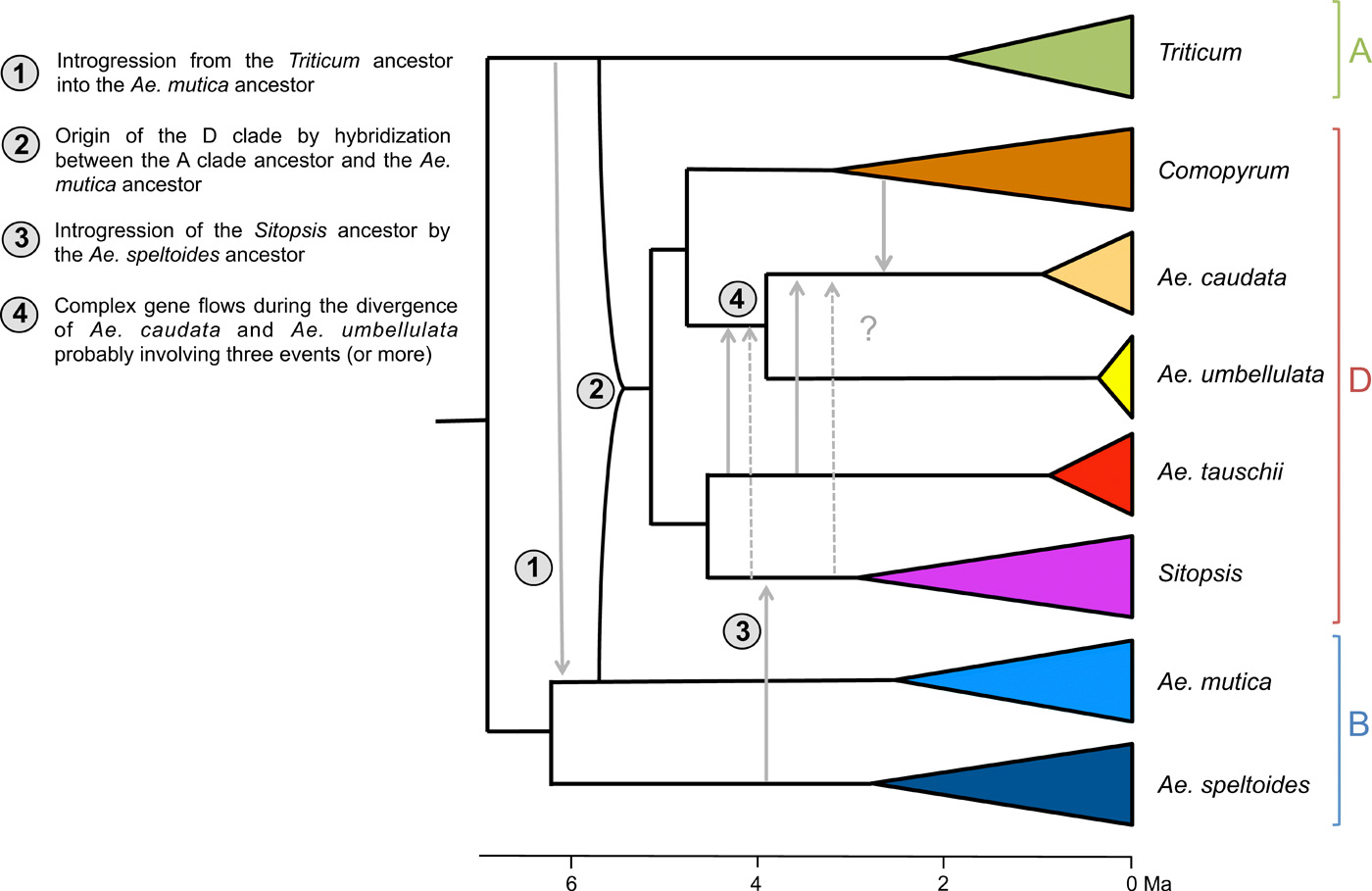
This is all that the world currently knows about the big empirical dataset (image source: https://advances.sciencemag.org/content/advances/5/5/eaav9188/F5.large.jpg?width=800&height=600&carousel=1):