Empirical dataset
We want to run NetRAX on this empirical dataset (from https://advances.sciencemag.org/content/5/5/eaav9188) which is available for download at https://bioweb.supagro.inra.fr/WheatRelativeHistory/index.php?menu=download
- We need the IndividualAlignments data, which we merge together into one big MSA (I wrote a script for it here: https://github.com/lutteropp/NetRAX/blob/master/scripts/merge_gene_alignments.py)
- Each gene is its own partition.
- The standard RAxML model (GTR+GAMMA) is okay
- We have 47 individuals over 17 species in the dataset.
- There are 1387815 patterns in the merged MSA, and 8738 partitions
- In order to create a MSA based on species, we can either use extended DNA alphabet or, more elegant version: Chapter 5 of Alexeys PhD thesis (https://cme.h-its.org/exelixis/pubs/dissAlexey.pdf)
That's the tree inferred by raxml-ng, which I will use as start network for NetRAX:
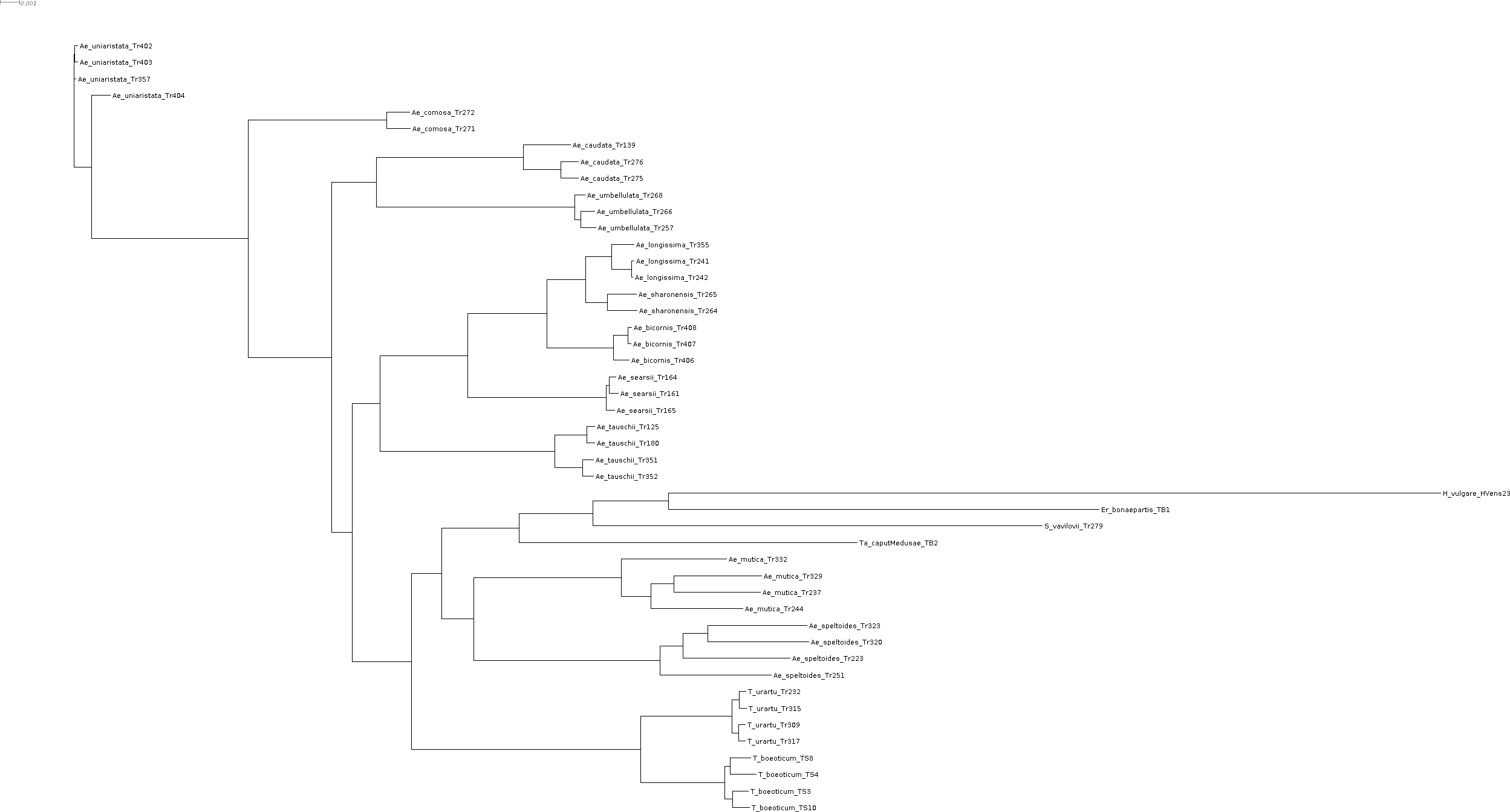 merged_genes.raxml.bestTree.txt
merged_genes.raxml.bestTree.txt
raxml-ng vanilla (with 10 random and 10 parsimony starting trees, site repeats and coarse-grain load balancing disabled, taking binary MSA file as input, on 20 cluster nodes) on the big empirical dataset needs about 4 hours in total, which is 15 minutes per starting tree. I used the master branch of raxml-ng for tree inference, commit 6db1154d64b5b6f65c09dccdc4576a8e5ab87195.
The exact call was:
/home/luttersh/raxml-ng/bin/raxml-ng-mpi --search --msa /home/luttersh/NetRAX/simulator/src/network/logic/data/datasets_big_empirical/merged_genes_msa.txt.raxml.rba --seed 42 --prefix /home/luttersh/NetRAX/simulator/src/network/logic/data/datasets_big_empirical --redo --site-repeats off --workers 1
This is the best 3-reticulation network we found so far, with running NetRAX under the Likelihood.BEST linked brlens model on the complete empirical dataset, using the RAxML-NG bestTree as single starting network (the inference is still running):
IMPROVED GLOBAL BEST SCORE FOUND SO FAR (3 reticulations): 60243776.78
((((Ae_uniaristata_Tr402:0.000125784,Ae_uniaristata_Tr403:0.000123531):4.10363e-05,Ae_uniaristata_Tr357:9.93126e-05):0.000845551,((Ae_comosa_Tr272:0.00111449,Ae_comosa_Tr271:0.00113325):0.00661148,((((((Ae_tauschii_Tr125:0.000360638,Ae_tauschii_Tr180:0.000374613):0.00152522,(Ae_tauschii_Tr351:0.000500896,Ae_tauschii_Tr352:0.000506094):0.0013587):1e-06)#2:0.00369354::0.699768,((((Ae_searsii_Tr164:0.000317254,Ae_searsii_Tr161:0.000435423):0.000162428,Ae_searsii_Tr165:0.00039857):0.00481951,((((Ae_longissima_Tr355:0.00105607,(Ae_longissima_Tr241:0.000105223,Ae_longissima_Tr242:7.27237e-05):0.000956713):0.00126074,(Ae_sharonensis_Tr265:0.0013944,Ae_sharonensis_Tr264:0.0013974):0.00103572):0.00191245,((Ae_bicornis_Tr408:0.000150353,Ae_bicornis_Tr407:0.000141744):0.000708389,Ae_bicornis_Tr406:0.000768194):0.00316566):1e-06)#1:0.00181396::0.78021):6.84414e-05)#0:0.00234715::0.696812):0.00229962,((Ae_caudata_Tr139:0.00222569,(Ae_caudata_Tr276:0.000819952,Ae_caudata_Tr275:0.000830589):0.00182064):0.00694046,(Ae_umbellulata_Tr268:0.000496678,(Ae_umbellulata_Tr266:0.000678434,Ae_umbellulata_Tr257:0.00075121):0.000294658):0.00944113):0.0022649):0.00105285,((((((H_vulgare_HVens23:0.0366505,Er_bonaepartis_TB1:0.0203093):0.00356795,S_vavilovii_Tr279:0.0210451):0.00367883,Ta_caputMedusae_TB2:0.0160405):0.00373834,((Ae_mutica_Tr332:0.00494289,((Ae_mutica_Tr329:0.00409685,Ae_mutica_Tr237:0.00408691):0.00114012,Ae_mutica_Tr244:0.00431241):0.00145895):0.00712706,(((((Ae_speltoides_Tr323:0.00464044,Ae_speltoides_Tr320:0.00476513):0.00122376,Ae_speltoides_Tr223:0.00485717):0.0011341,Ae_speltoides_Tr251:0.00527979):0.00447101,#0:0.00626098::0.303188):0.00223551,#2:0.011478::0.300232):0.00223551):0.00156377):0.00153908,(((T_urartu_Tr232:0.000340193,T_urartu_Tr315:0.000345844):0.000339418,(T_urartu_Tr309:0.000310887,T_urartu_Tr317:0.000326565):0.000320805):0.004184,((T_boeoticum_TS8:0.000895566,T_boeoticum_TS4:0.00113021):0.000240495,(T_boeoticum_TS3:0.00067409,T_boeoticum_TS10:0.000717175):0.000342407):0.00365916):0.0112862):0.00213079,#1:0.00858681::0.21979):0.00117325):0.00402145):0.00744299):0.000844198,Ae_uniaristata_Tr404:3.8904e-05);
This is all that the world currently knows about the big empirical dataset (image source: https://advances.sciencemag.org/content/advances/5/5/eaav9188/F5.large.jpg?width=800&height=600&carousel=1):
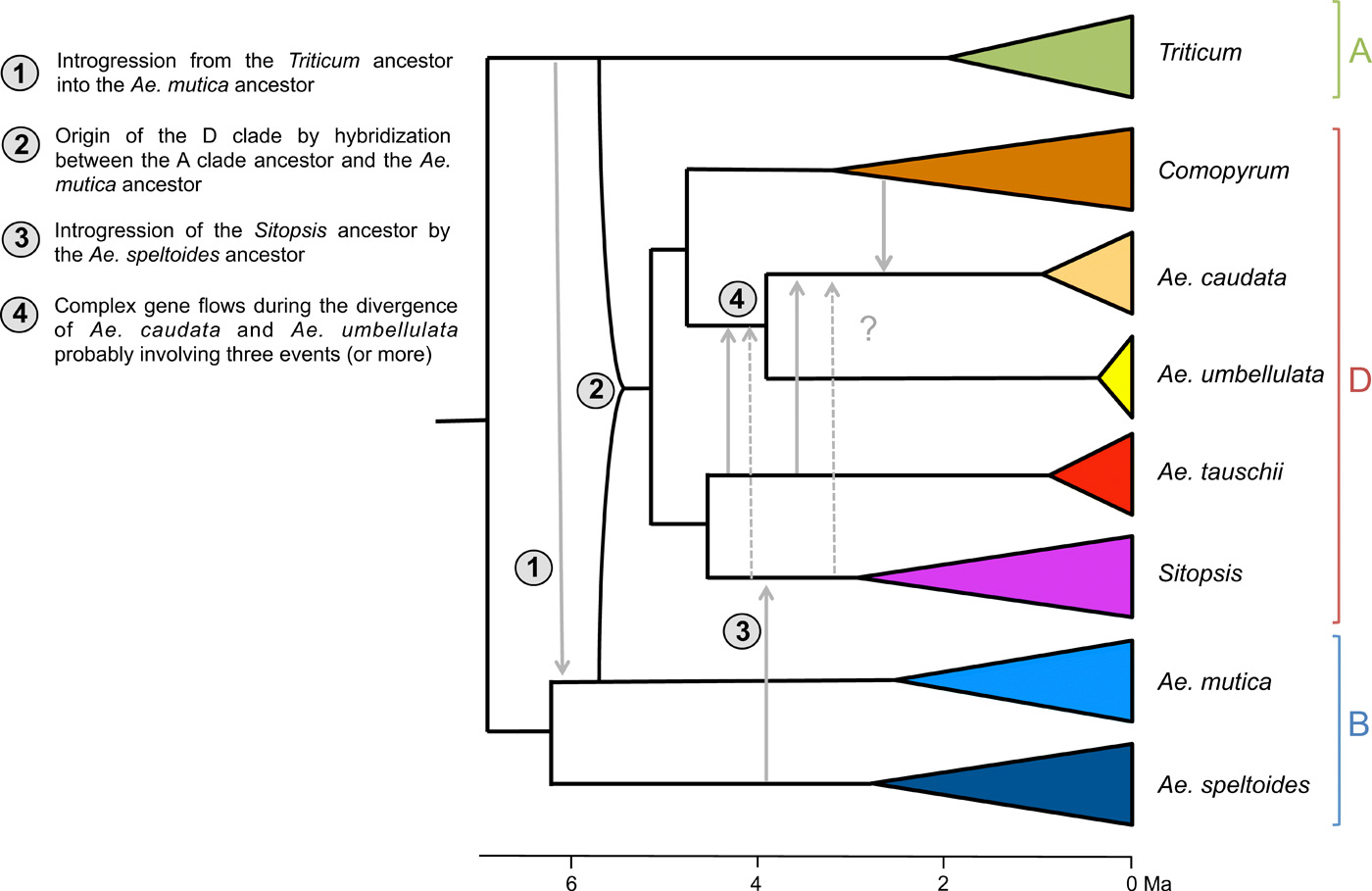
I am confused why there are only 8 groups in this image, because from the single-gene alignments we end up with 17 species according to my data merging Python script:
{'Ae_bicornis', 'Ae_speltoides', 'Ae_mutica', 'Ta_caputMedusae', 'T_urartu', 'H_vulgare', 'Ae_tauschii', 'Er_bonaepartis', 'Ae_caudata', 'T_boeoticum', 'Ae_umbellulata', 'Ae_longissima', 'Ae_uniaristata', 'Ae_sharonensis', 'Ae_searsii', 'S_vavilovii', 'Ae_comosa'}
I subsampled the empirical dataset, using majority consensus for the 17 species sequences.
That's the tree inferred by raxml-ng, which I will use as start network for NetRAX:
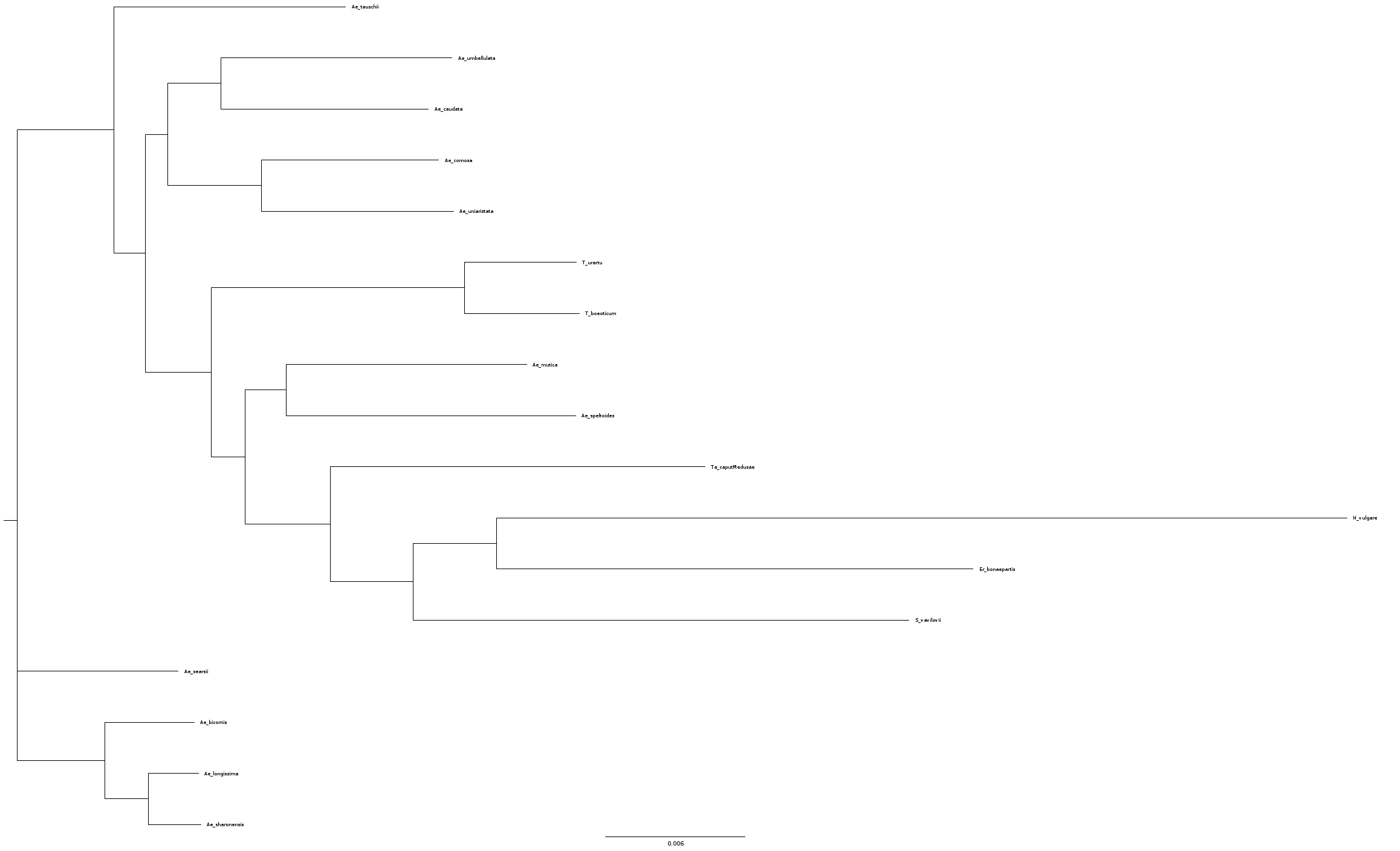
merged_genes_species.raxml.bestTree.txt
raxml-ng vanilla (with 10 random and 10 parsimony starting trees, site repeats and coarse-grain load balancing disabled, taking binary MSA file as input, on 10 cluster nodes) on the big empirical dataset needs about 1 hour in total, which is around 3 minutes per starting tree. I used the master branch of raxml-ng for tree inference, commit 6db1154d64b5b6f65c09dccdc4576a8e5ab87195.