vincentappiah
vincentappiah
Hi, I also have a similar issue. Especially the image attached 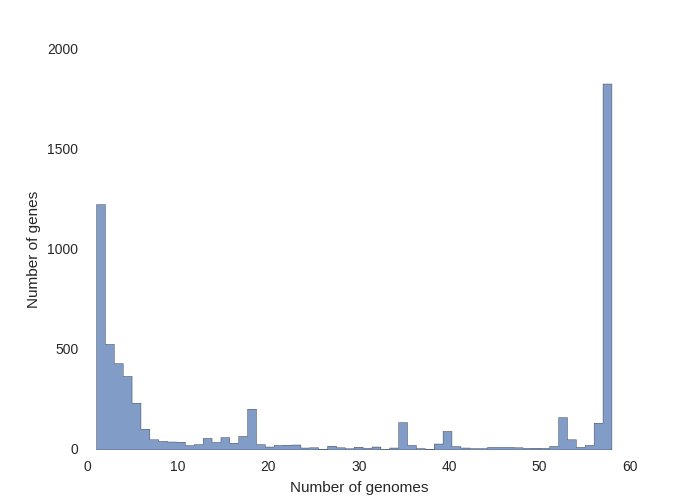 For eg. if lets say the number of genomes is 10. which of the 60 genomes does it...
Dear @felipelira They are fragmented genes.
@atks I had a similar issue. Below is is my error [variant_manip.cpp:96 is_not_ref_consistent] reference bases not consistent: chr10:4868478-4868486 TGCGGGGCG(REF) vs Tgcggggcg(FASTA) When I used the -n flag vt skipped several...
I guess I can still use the normalized vcf. I can send you the original VCF in case you want to examine it.
Hi @pd3 Here is the output for the grep command. chr8 **160826** . C T 1139.53 . DPB=73;EPPR=5.63551;GTI=0;MQMR=60;NS=1;NUMALT=1;ODDS=80.0471;PAIREDR=1;PQR=0;PRO=0;QR=1847;RO=67;RPPR=5.63551;SRF=16;SRP=42.7126;SRR=51;DP=124;AB=0;ABP=0;AF=1;**AO=44**;CIGAR=3M1X;DPRA=0;EPP=12.6832;LEN=1;MEANALT=5;MQM=60;PAIRED=1;PAO=1;PQA=29;QA=1434;RPL=8;RPP=41.702;RPR=36;RUN=1;SAF=7;SAP=47.4268;SAR=37;TYPE=snp;technology.Illumina=1;AN=4;AC=2GT:DP:AD:RO:QR:AO:QA:GL 0/0:73:67,.:67:1847:.:.:0,.,. 1/1:51:1,44:1:39:44:1434:-125.592,-10.0141,0 ./.:.:.:.:.:::. chr8 **160826** . C T 1783.34 . DPB=69.6667;EPPR=5.18177;GTI=0;MQMR=60;NS=1;NUMALT=1;ODDS=95.5437;PAIREDR=1;PQR=57;PRO=2;QR=2;RO=1;RPPR=5.18177;SRF=0;SRP=5.18177;SRR=1;DP=67;AB=0;ABP=0;AF=1;**AO=66**;CIGAR=2M1X;DPRA=0;EPP=3.53672;LEN=1;MEANALT=1;MQM=60;PAIRED=1;PAO=3;PQA=91;QA=2097;RPL=27;RPP=7.74806;RPR=39;RUN=1;SAF=10;SAP=72.629;SAR=56;TYPE=snp;technology.Illumina=1;AN=2;AC=2GT:DP:AD:RO:QR:AO:QA:GL 1/1:67:1,66:1:2:66:2097:-188.514,-20.2481,0...
Hello All, I am working an Mycobacterium ulcerans genome which was sequenced with oxford nanopore technology. I am trying to do denovo assembly with flye but I run into a...
Please is there a way to know the expected genome size before hand?
@fenderglass is there a way to know the expected genome size before starting the assembly?
@eyad. This is what worked for me I looked up the genome size of my organism (in my case 6.5mb)In the flye software, Flye still raised the flag. I reduced...
Thanks. I have a bacterial sequence (M. ulclerans) which I am annotating. Pseudogene was detected _/note="PseudoGeneDetection:WP_013225067.1 formate_ Is there anyway for DFAST to give how many of the pseudogenes are...