lokapal
lokapal
As a matter of fact I have FaeI sequence (palindrome CATG) AFTER that I need to remove all to the end of the read, but retain FaeI itself (it's common...
It doesn't work at all, these sequence were not trimmed at all (I guess retain is in effect): @HISEQ:XXXXXXXXXXXXXX 1:N:0:TATAATAT GAATTCTTCACTGGGGCTCCTAGGATGGGCTGGTTGGGTGGGGGTGGAGAATGTCTGCCTCCAGAGTTTGCATGGA @HISEQ:XXXXXXXXXXXXXX 1:N:0:TATAATAT TGTGACCAAAAGTGCCTCTGCTGGGAGCTGGGATCCTCGGGACCATGATCCCAACAATGTTCACAG The commandline was: `cutadapt -j 20...
I've tried to build linked adapter something like to: `"CATG;required;action=retain;noindels;min_overlap=4;max_error_rate=0.01...N{20}X;optional;action=trim"` but to no avail: action per adapter is not supported
As a matter of fact my current reads have THREE adapters: one of them is full, the second is broken (sometimes at the head with the end presenting, sometimes at...
Surely I do, it's not a theoretical question. Please find attached the example: two entries that are marked up. It's 4C library. [reads.fa.gz](https://github.com/jdidion/atropos/files/6949545/reads.fa.gz) Three adapters: A1, A2, Illumina/IlluminaPE. A1D, A2D...
I can't state for "all" researchers but "my" wet biologists constantly supply me with libraries that can contain two 4C adapters and can contain only one 4C adapter, can contain...
 ``` > print(sessionInfo()) R version 4.0.5 (2021-03-31) Platform: x86_64-w64-mingw32/x64 (64-bit) Running under: Windows 7 x64 (build 7601) Service Pack 1 Matrix products: default attached base packages: [1] grid stats...
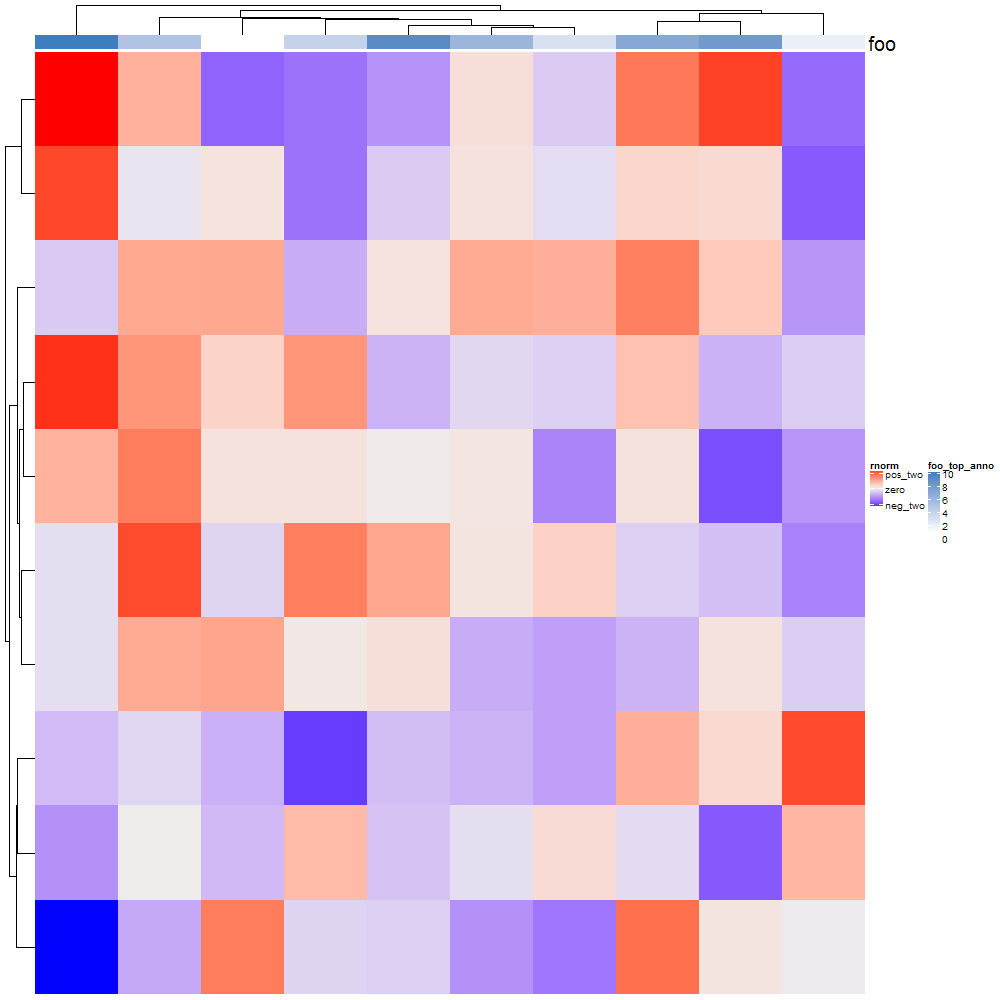
I cannot reverse this heatmap by any means. I've tried to change colors order: f1 = colorRamp2(c(-1.5, -2, -5),c("lightblue", "blue", "darkblue")) And I've tried with at = The result is...
What is your CPU? It works flawlessly at Intel CPUs, even old ones like Xeon 2697 v2. My issue is concerning modern AMD Ryzens (5xxx series).
Well, I prefer to keep mine (and all other FFs that I install) to ESR versions. And it has a lot of sense.