lh12565
lh12565
Hi, I have a simple questions about rank acceleration. Is `abs` just changing the sign? If yes, Why is `BIK` ahead of `S100A6` in column 1 of `rank_abs_acceleration` as follows:...
Hi, my data was obtained the conventional scRNA-seq technique (10X) with no labeling data. I found the nonsensical velocity flow from macrophage to monocyte using dynamo. I found your article...
Hi, I try to install molgrid using conda-molgrid. When I finished installing molgrid, there was an error installing RDKit: ``` conda install -c conda-forge rdkit Solving environment: failed with repodata...
Hi, I try to install dyno in windows. An error has occurred: ```R devtools::install_github("dynverse/dyno") ``` ``` C:/Rtools/mingw_64/bin/../lib/gcc/x86_64-w64-mingw32/4.9.3/../../../../x86_64-w64-mingw32/bin/ld.exe: skipping incompatible C:/PROGRA~1/R/R-3.6.2/bin/i386/R.dll when searching for -lR C:/Rtools/mingw_64/bin/../lib/gcc/x86_64-w64-mingw32/4.9.3/../../../../x86_64-w64-mingw32/bin/ld.exe: skipping incompatible C:/PROGRA~1/R/R-3.6.2/bin/i386/R.dll when searching...
Hi, @mattragoza I installed LiGAN successfully. But when I run pytest tests, there is a lot of errors occurred: ``` ............. _ _ _ _ _ _ _ _ _...
Hi, I use 'prepare_ligand4.py' to convert small molecules from mol2 to pdbqt. I found some molecules changed their structures after converting to pdbqt, for example: 1.mol2: 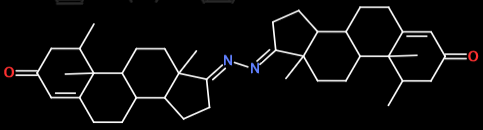 after convert (1.pdbqt):...
Hi, my system is CentOS7. when I install the coordgen, there is an error as follows: ``` make -j install [ 85%] Building CXX object example_dir/CMakeFiles/example.dir/example.cpp.o [ 90%] Linking CXX...
Hi, I have a json string not a json file. How can I resolve the json string? There is an error if run the code as follows: ```javascript d3.queue() .defer(d3.json,...
Hi when I run browserify, there is a error as below: ``` browserify -t browserify-css -t svgify ../test/node_modules/react-mutation-mapper/dist/index.js -o all.js /NAS/lh/down/npm_Data/browserify/node_modules/buffers/index.js:185 return this.buffers[pos.buf].get(pos.offset); ^ TypeError: this.buffers[pos.buf].get is not a function...
Hi, I use funcotator to call somatic mutations as below: ``` gatk Funcotator \ -R $ref/Homo_sapiens_assembly38.fasta \ -V ../${array[1]}.filter.vcf \ -O ${array[1]}.maf \ --output-file-format MAF \ --data-sources-path Funcotator_data/funcotator_dataSources.v1.7.20200521s/ \ --ref-version...
