Hanbin Lee
Hanbin Lee
I've been trying to understand how splicing works on ondisk storage. I currently have a csc sparse matrix where row are genes (~30000) and column are cells (~2 million). Suppose...
Hi there, thank you for developing a terrific library. I'm trying to perform certain matrix operations using tskit. Suppose that A is a matrix in which the number of rows...
Hi, I found out something strange when using batch correction. These are the two result from Muraro (pancreas) and MacParland (liver) datasets. This is from Muraro 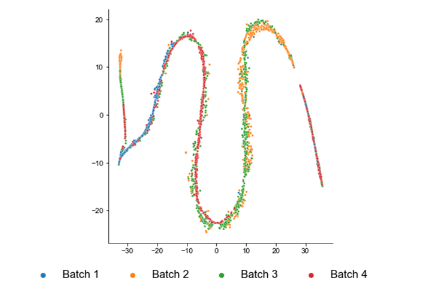 and this is...
## Description A draft of randomized principal component analysis (PCA) using the `TreeSequence.genetic_relatedness_vector`. The implementation contains `spicy.sparse` which should eventually be removed. This part of the code is only used...
A `ZeroDivisionError` happens in the following example (tsdate version 0.2.1) ``` import tskit import msprime import tsinfer import tsdate # simulate tree sequence demography = msprime.Demography() demography.add_population(name="Panmictic", initial_size=100_000, growth_rate=0.01) num_individuals...
Continuing from https://github.com/tskit-dev/tskit/pull/3077. I think this link from numpy docs is a good starting point: https://numpy.org/doc/2.2/reference/random/multithreading.html The bottom line is that one can execute multiple computations concurrently with `concurrent.futures.ThreadPoolExecutor` given...
