Zach
Zach
Additionally, these maps do not differ when viewed simultaneously in WashU Epigenome Browser: 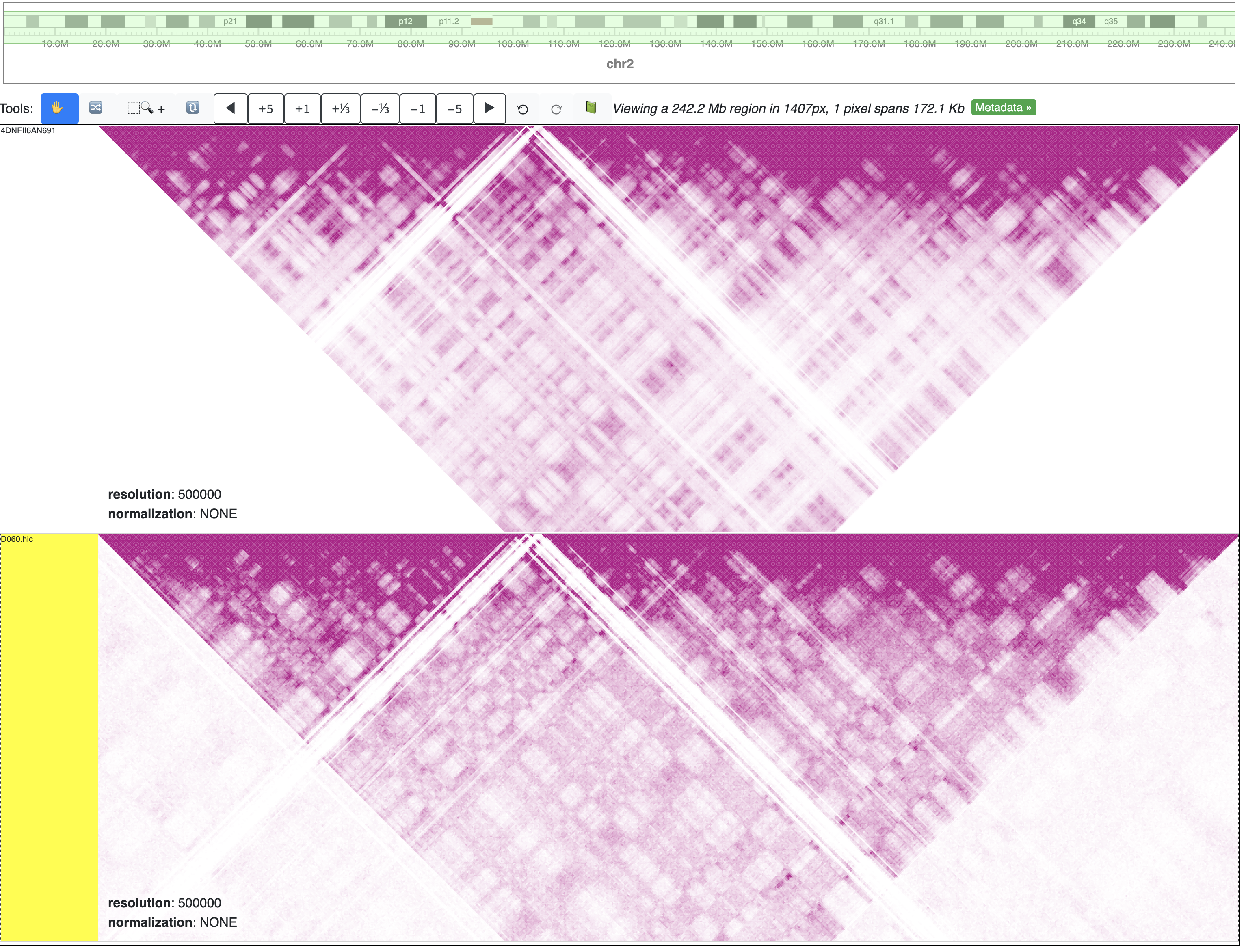
As I stated, the issue is exactly the same in the desktop version. Local D60 file loaded alone on desktop version: 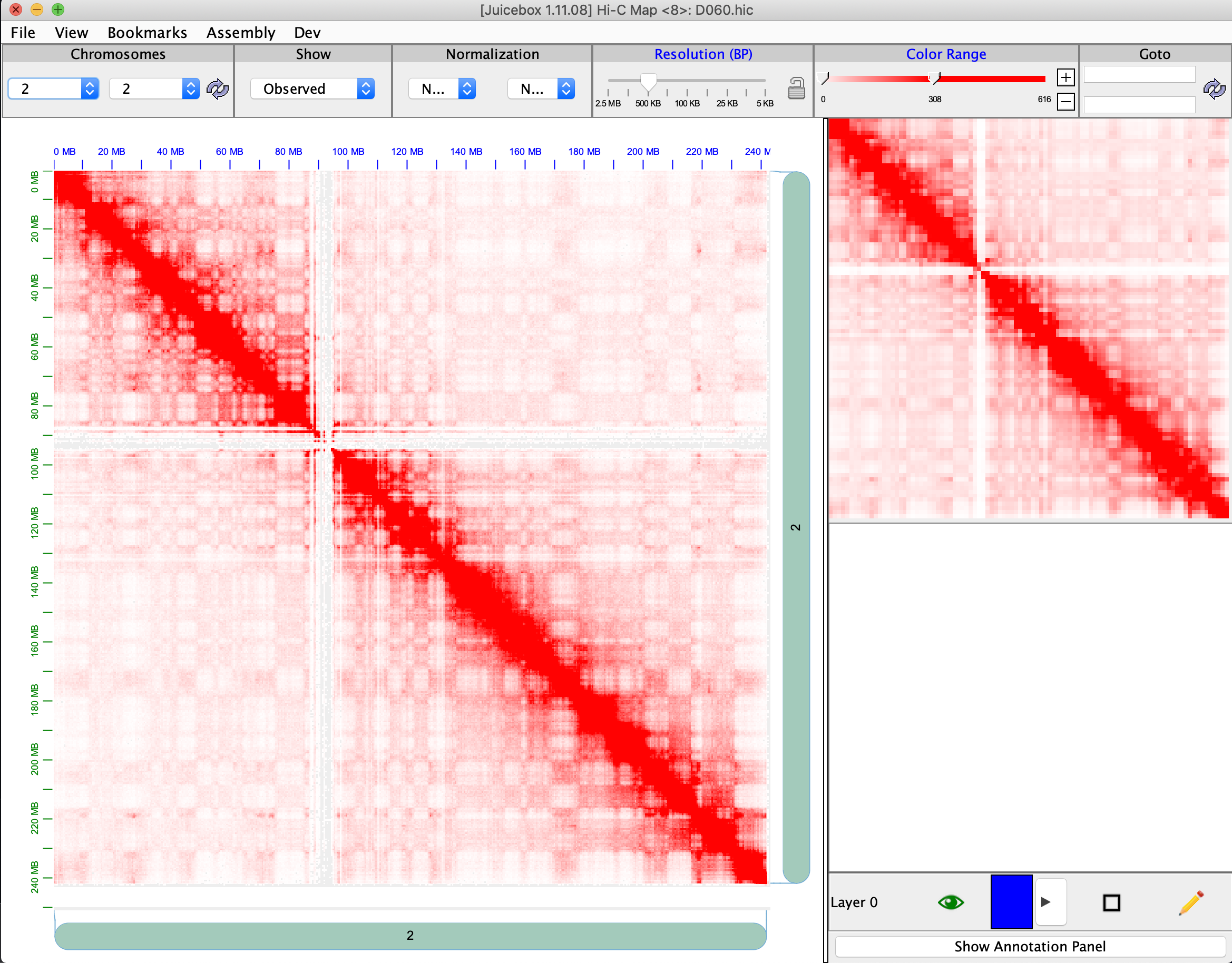 4DN file loaded alone on desktop version: 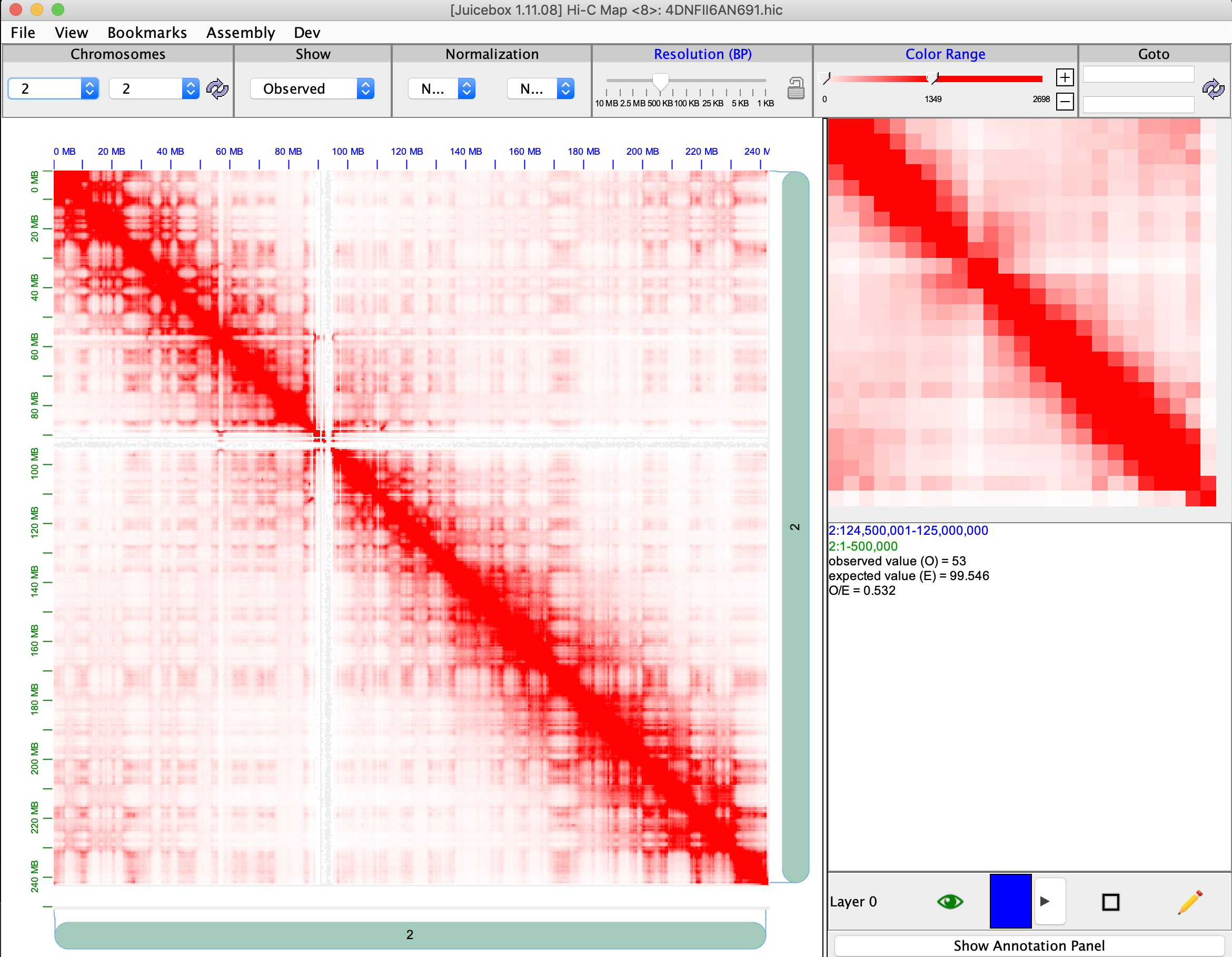...
No worries. D60 validation output: ``` $ juicer_tools validate ../data/020_final_output/hic/D060.hic Reading file: ../data/020_final_output/hic/D060.hic File has normalization: VC Description: Coverage File has normalization: VC_SQRT Description: Coverage (Sqrt) File has normalization: KR...
Just echo-ing this -- I got the exact same error running the nf-core/cutandrun v3.2-g506a325 version of the pipeline on Nextflow 23.04.3 in Slurm/HPC setting.
Sure, I actually just had some C&R data come off the sequencer on Friday so I planned to run the pipeline this week anyways. I'll follow up in the next...