losses went into inf/nan if optimizer is not SGD
I tested other optimizers like adam/rmsprop/momentum from 'jax.example_libraries.optimizers'. As design initiated, the whole structure predicted by AlphaFold2 became a “black hole” after 10-20 steps, then loss value changed into inf/nan after 60-80 steps, yet the inputs seems normal. I tried several times per optimizer and got the same result.
Oops. I fixed this issue in the beta branch, and just pushed the fix to the main branch.
Can you try again?
Thank you sir, that was fast. Still the "black hole" bug exists(especially in the soft_iters, there is a sharp decline and then a sharp increase in losses), seems Evofomer learnt chaos from self._grad & self._state that cannot provide meaningful information to structure module.
Hi Peldom, can you share the example you are working with? I'll investigate.
key codes: (adam, rmsprop, etc)
protocol == "binder"
from jax.example_libraries.optimizers import rmsprop
model._setup_optimizer(optimizer=rmsprop)
model.design_3stage(soft_iters=40, temp_iters=40, hard_iters=20)
And the structure is predicted like this when all the losses are quiet low(around step 20)
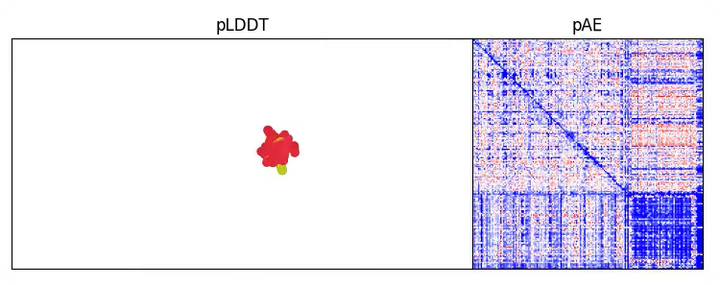 Take 'pae_intra' for example:
Take 'pae_intra' for example:
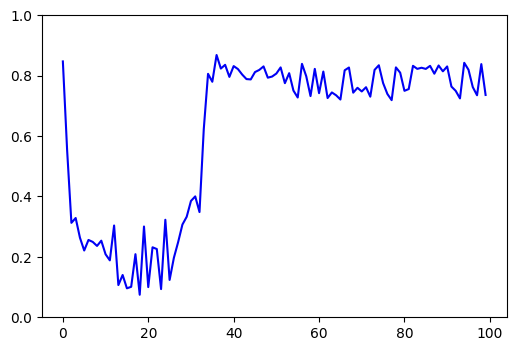
I'm so confused😅 , it happens more often in soft_iters than temp_iters/hard_iters, I would be so grateful if you could find out why
Did you change how the sequence was initialized? Or is this the result of changing the optimizer?
The current optimizer was calibrated (zero initialization, dropouts, switching between model params, normalized gradients) to avoid these adversarial modes.
Can you share the example you are trying on? (if it's private, you can email me at [email protected]).