How to ignore values outside the mask when calculating `distance`
Hi,
I am calculating the distance from non-forest cells to the closest forest cell in a raster of forests in a municipality. Below is the code to reproduce it.
# packages
library(dplyr)
library(sf)
library(terra)
library(geobr) # for downloading vector of the study area
# options
options(timeout = 600)
# download raster
download.file(url = "https://storage.googleapis.com/mapbiomas-public/brasil/collection-6/lclu/coverage/brasil_coverage_2020.tif",
destfile = "brasil_coverage_2020.tif", mode = "wb")
# import raster
mapbiomas <- terra::rast("brasil_coverage_2020.tif")
mapbiomas
# rio claro municipality - vector
rc <- geobr::read_municipality(code_muni = 3543907, year = 2020) %>%
sf::st_transform(4326) %>%
terra::vect()
rc
# plot
# plot(mapbiomas, col = viridis::viridis(20))
# plot(rc, col = "red", add = TRUE)
# crop and mask
mapbiomas_rc <- mapbiomas %>%
terra::crop(rc) %>%
terra::mask(rc)
# forest
forest <- mapbiomas_rc == 3
plot(forest)
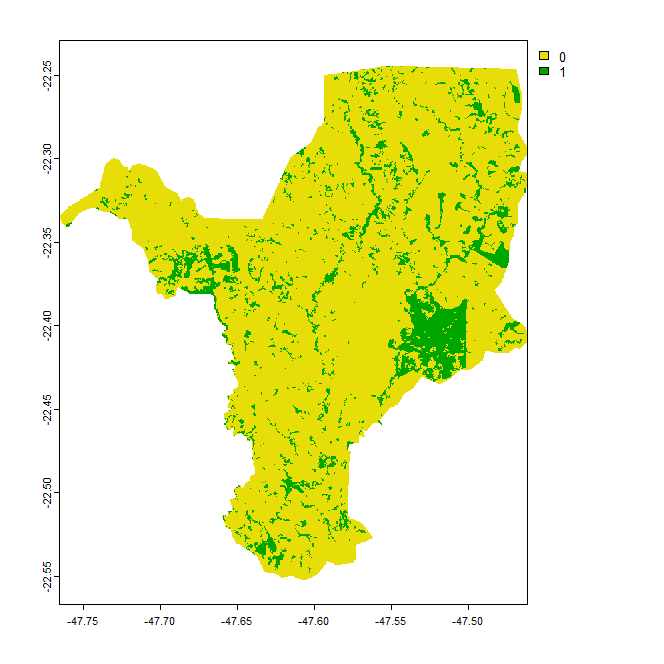
Here is the plot of the forest areas (class 3 in the original land use map, reclassified as 1) against non-forest areas (reclassified as 0) after masking out all pixels outside the municipality I am interested in (these pixels are now NA outside the mask). I am interested in calculating the distance of all non-forest pixels (class 0) to the nearest forest pixel (class 1). To do so, I need to transform the class 0 into NA, since the terra::distance() function works like this.
# forest na
forest_na <- forest
values(forest_na) <- ifelse(values(forest_na) == 1, 1, NA)
plot(forest_na)
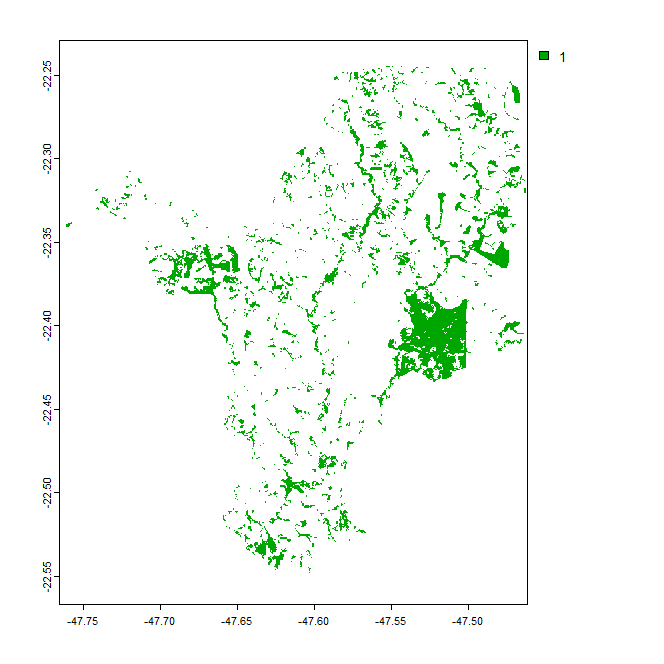
The last command produces a map with 1 for forest areas and NA for non-forest areas. There is no difference, however, between the NA cells that represent non-forest within the municipality limits (that were 0 in the first map) and the NA cells masked out - so that terra::distance() tries to calculate the distance from all those NA pixels (even those masked out) to the closest non-NULL and this takes ages - it di not run in my personal computer.
Is there a way to really mask out - i.e. disconsider these NA pixels that were masked out in the calculation - so that the procedure is performed only for NA cells within the mask? In any case, it seems this process is time consuming, at least for a landscape of this size - 1144 rows per 1126 cols.
If that is really the code you are using, consider saving all intermediate results to file (with filename = "" at each terra function) to free up memory. perhaps that will help. Also use classify( filename = "") instead of ifelse
Hi @DjanerEmin thanks for the suggestions. The map is not big, though, so this should not be a problem of memory, I believe.
Thank you for the suggestion. It is now possible to ignore cells. I am very sorry it took me so long to respond.
library(terra)
library(geobr)
options(timeout = 600)
url = "https://storage.googleapis.com/mapbiomas-public/brasil/collection-6/lclu/coverage/brasil_coverage_2020.tif"
fname <- basename(url)
if (!file.exists(fname)) { download.file(url, fname, mode = "wb") }
mapbiomas <- terra::rast("brasil_coverage_2020.tif")
rc <- vect(geobr::read_municipality(code_muni = 3543907, year = 2020))
rc <- project(rc, "epsg:4326")
mapbiomas_rc <- mapbiomas |> terra::crop(rc, mask=T)
forest <- mapbiomas_rc == 3
## aggregating to speed up the example
aforest <- aggregate(forest, 10, "modal")
aforest <- aggregate(forest, 10, "modal")
d = distance(aforest, target=0)
plot(d)
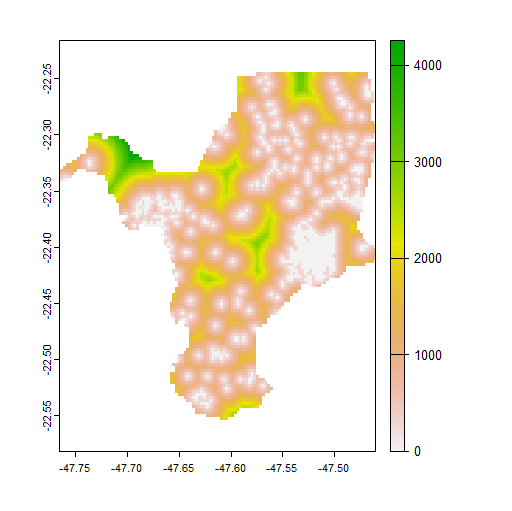
This can indeed save a lot of time; but distance is slower than it should be and it is another goal to improve that.
Great, thanks, @rhijmans !