*S(=O)* stereochemistry differences between OE and RD
Describe the bug
I used off_coverage.py to cross-compare the OpenFF Molecule objects created by OpenEyeToolkitWrapper with those created by RDKitToolkitWrapper, using oe_mol.is_isomorphic_with(rd_mol). The failure cases were only those with a chiral *-S(=O)-*. The underlying toolkits handle this chirality differently.
This is related to https://github.com/openforcefield/openff-toolkit/issues/146 (see the entry containing CCS(=O)C) and https://github.com/openforcefield/openff-toolkit/issues/467 where the molecules on the right, with the *-S(=O)-* "cannot be loaded without allow_undefined_stereo=True".
To Reproduce
With the off_coverage tool (about to be merged), I'll have it process MiniDrugBank.sdf and list the identifiers where the two topologies are not isomorphic (I'll use the description file to show that's what xcmp_isomorphic_err means.)
% python off_coverage.py xcompare MiniDrugBank.sdf -o mdb.feats --description mdb.description
.. output omitted ..
% grep "are not isomorphic" mdb.description
xcmp_isomorphic_err OE and RD topologies are not isomorphic
% grep xcmp_isomorphic_err mdb.feats | awk '{printf("%s ", $1)} END {print ""}'
DrugBank_3817 DrugBank_4032 DrugBank_1971 DrugBank_2140 DrugBank_2563 DrugBank_2585 DrugBank_2687
For something a little more direct:
import pathlib
from openff import toolkit
from openff.toolkit.utils import toolkits
from io import BytesIO
oe_wrapper = toolkits.OpenEyeToolkitWrapper()
rd_wrapper = toolkits.RDKitToolkitWrapper()
ids = "DrugBank_3817 DrugBank_4032 DrugBank_1971 DrugBank_2140 DrugBank_2563 DrugBank_2585 DrugBank_2687".split()
filename = pathlib.Path(toolkit.__file__).parent / "data" / "molecules" / "MiniDrugBank.sdf"
content = filename.read_bytes()
for id in ids:
i = content.find(id.encode("utf8"))
j = content.find(b"$$$$\n", i) + 5
record = content[i:j]
oe_mol = oe_wrapper.from_file_obj(BytesIO(record), "sdf")[0]
rd_mol = rd_wrapper.from_file_obj(BytesIO(record), "sdf")[0]
print(id, oe_mol.is_isomorphic_with(rd_mol))
print(" OpenEye:", oe_wrapper.to_smiles(oe_mol))
print(" RDKit:", rd_wrapper.to_smiles(rd_mol))
Output
The output from the above program, including warning message from OEChem, is:
Warning: Invalid double bond stereomark ignored on bond number 42 of DrugBank_3817
DrugBank_3817 False
OpenEye: [H]/C(=C(/[H])\C(=O)N([H])[C@@]([H])(C([H])([H])O[H])C([H])([H])[S@@](=O)C([H])([H])SC([H])([H])[H])/C1=C(N(C(=O)N(C1=O)[H])[H])C([H])([H])[H]
RDKit: [H][O][C]([H])([H])[C@]([H])([N]([H])[C](=[O])/[C]([H])=[C](\[H])[C]1=[C]([C]([H])([H])[H])[N]([H])[C](=[O])[N]([H])[C]1=[O])[C]([H])([H])[S@@](=[O])[C]([H])([H])[S][C]([H])([H])[H]
Warning: Invalid double bond stereomark ignored on bond number 38 of DrugBank_4032
DrugBank_4032 False
OpenEye: [H]c1c(c(c(c(c1[C@]([H])(C([H])([H])[H])N([H])C(=O)[C@@]2([C@](C2(Cl)Cl)([H])C([H])([H])[H])[S@@](=O)C([H])([H])[H])[H])[H])Br)[H]
RDKit: [H][c]1[c]([H])[c]([C@@]([H])([N]([H])[C](=[O])[C@@]2([S@@](=[O])[C]([H])([H])[H])[C]([Cl])([Cl])[C@@]2([H])[C]([H])([H])[H])[C]([H])([H])[H])[c]([H])[c]([H])[c]1[Br]
Warning: Invalid double bond stereomark ignored on bond number 20 of DrugBank_1971
DrugBank_1971 False
OpenEye: [H][C@@](C(=O)O[H])(C([H])([H])C([H])([H])[S@](=O)C([H])([H])[H])N([H])[H]
RDKit: [H][O][C](=[O])[C@@]([H])([N]([H])[H])[C]([H])([H])[C]([H])([H])[S@](=[O])[C]([H])([H])[H]
Warning: Invalid double bond stereomark ignored on bond number 34 of DrugBank_2140
DrugBank_2140 False
OpenEye: [H]C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[S@@](=O)C([H])([H])C([H])([H])O[H]
RDKit: [H][O][C]([H])([H])[C]([H])([H])[S@](=[O])[C]([H])([H])[C]([H])([H])[C]([H])([H])[C]([H])([H])[C]([H])([H])[C]([H])([H])[C]([H])([H])[C]([H])([H])[H]
Warning: Invalid double bond stereomark ignored on bond number 26 of DrugBank_2563
DrugBank_2563 False
OpenEye: [H][C@@]1(C(C([S@@](=O)C1([H])[H])([H])[H])([H])[H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H]
RDKit: [H][C]([H])([H])[C]([H])([H])[C]([H])([H])[C]([H])([H])[C@]1([H])[C]([H])([H])[S@](=[O])[C]([H])([H])[C]1([H])[H]
Warning: Invalid double bond stereomark ignored on bond number 18 of DrugBank_2585
DrugBank_2585 False
OpenEye: [H]C(=C([H])C([H])([H])[S@@](=O)C([H])([H])/C(=C(\[H])/S[H])/[H])[H]
RDKit: [H][S]/[C]([H])=[C](\[H])[C]([H])([H])[S@](=[O])[C]([H])([H])[C]([H])=[C]([H])[H]
Warning: Invalid double bond stereomark ignored on bond number 46 of DrugBank_2687
DrugBank_2687 False
OpenEye: [H][C@@]1(C(=O)N([C@]1([H])S[H])[C@]([H])(C(=O)O[H])C([H])([H])[S@@](=O)C([H])([H])[H])N([H])C(=O)C([H])([H])C([H])([H])C([H])([H])[C@@]([H])(C(=O)O[H])N([H])[H]
RDKit: [H][O][C](=[O])[C@@]([H])([N]([H])[H])[C]([H])([H])[C]([H])([H])[C]([H])([H])[C](=[O])[N]([H])[C@@]1([H])[C](=[O])[N]([C@]([H])([C](=[O])[O][H])[C]([H])([H])[S@@](=[O])[C]([H])([H])[H])[C@]1([H])[S][H]
You'll notice the "Warning: Invalid double bond stereomark" occurs while processing each record.
This is because the V2000 molblock uses a bond stereo of "1" (as with DrugBank_2140) or "6" (as with DrugBank_2585 and DrugBank_2563), but the ctfile specification says that those values do not apply to double bonds.
I replaced those values with 0 and those warnings disappeared, but those three files were still not isomorphic. (I didn't check or test all 6 records).
If I replace the is_isomorphic_with() to ignore atom stereochemistry matching, using:
print(id, oe_mol.is_isomorphic_with(rd_mol, atom_stereochemistry_matching=False))
then all of these are isomorphic.
Further Analysis
Simple inspection of the SMILES output:
[H]/C(=C(/[H])\C(=O)N([H])[C@@]([H])(C([H])([H])O[H])C([H])([H])[S@@](=O)C([H])([H])SC([H])([H])[H])/C1=C(N(C(=O)N(C1=O)[H])[H])C([H])([H])[H] DrugBank_3817
[H]c1c(c(c(c(c1[C@]([H])(C([H])([H])[H])N([H])C(=O)[C@@]2([C@](C2(Cl)Cl)([H])C([H])([H])[H])[S@@](=O)C([H])([H])[H])[H])[H])Br)[H] DrugBank_4032
[H][C@@](C(=O)O[H])(C([H])([H])C([H])([H])[S@](=O)C([H])([H])[H])N([H])[H] DrugBank_1971
[H]C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[S@@](=O)C([H])([H])C([H])([H])O[H] DrugBank_2140
[H][C@@]1(C(C([S@@](=O)C1([H])[H])([H])[H])([H])[H])C([H])([H])C([H])([H])C([H])([H])C([H])([H])[H] DrugBank_2563
[H]C(=C([H])C([H])([H])[S@@](=O)C([H])([H])/C(=C(\[H])/S[H])/[H])[H] DrugBank_2585
[H][C@@]1(C(=O)N([C@]1([H])S[H])[C@]([H])(C(=O)O[H])C([H])([H])[S@@](=O)C([H])([H])[H])N([H])C(=O)C([H])([H])C([H])([H])C([H])([H])[C@@]([H])(C(=O)O[H])N([H])[H] DrugBank_2687
or the corresponding depiction (here with CDK Depict):
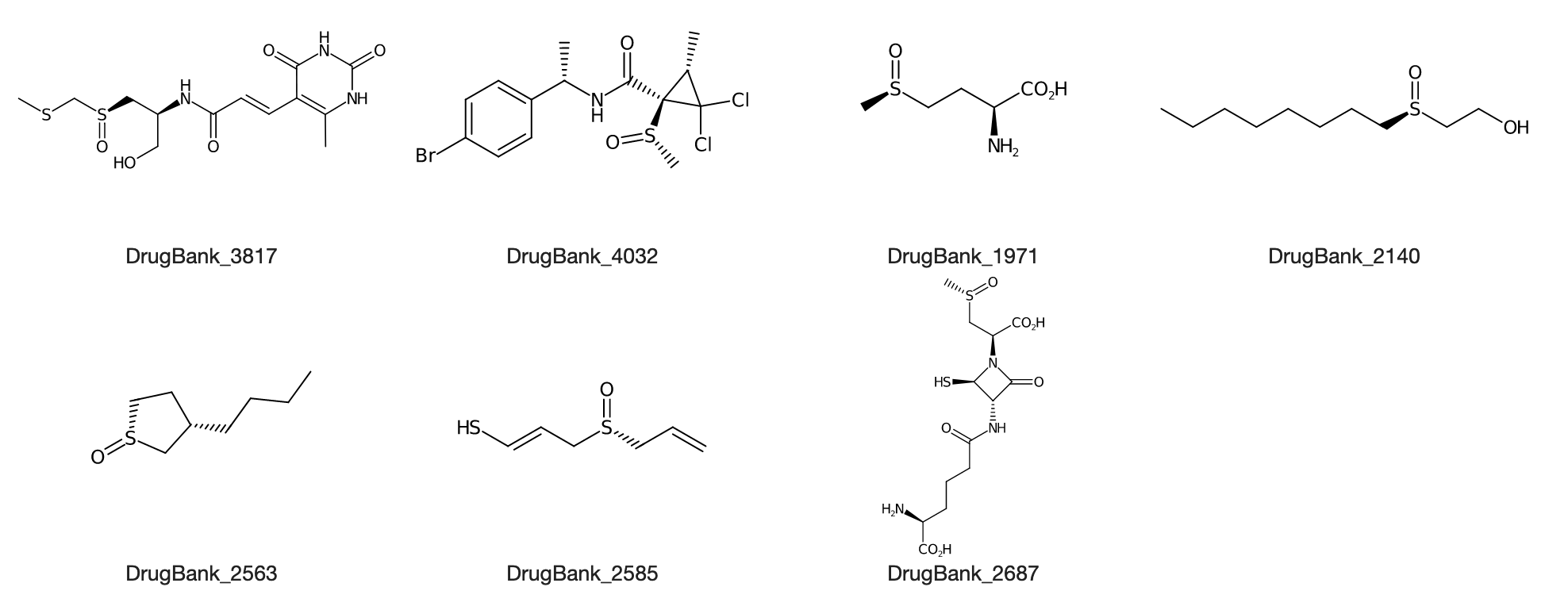
shows they all contain a chiral *-S(=O)-*