BioCircos.R

BioCircos.R copied to clipboard
How to change the thickness of chromosome track?
@koalive I really like BioCircos for its ease of use. Is there any way to change the height of chromosome track in R -- i refer to the green bar on chr12. I wanted to make it smaller. Thanks so much.
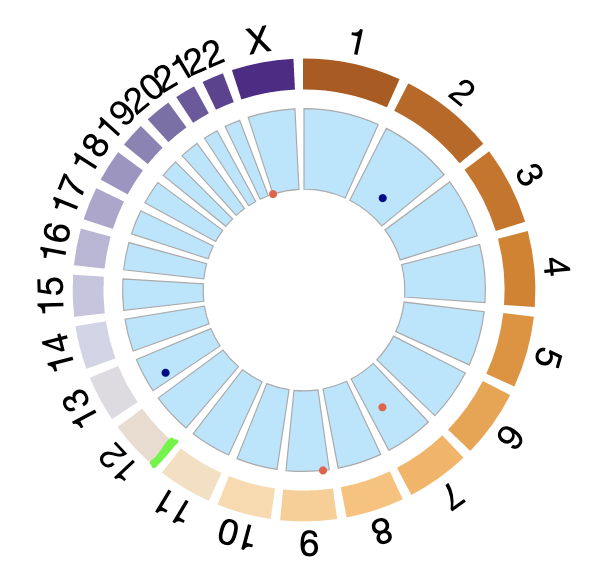
Hi. So you want to change the thickness of the genome track? So far there's no direct option for that but if you really need it you can always hide it (e.g. by making it white) and replacing it by a Background track or an Arc track:
library(BioCircos)
# Chromosomes on which the points should be displayed
points_chromosomes = c('X', '2', '7', '13', '9')
# Coordinates on which the points should be displayed
points_coordinates = c(102621, 140253678, 98567307, 28937403, 20484611)
# Values associated with each point, used as radial coordinate
# on a scale going to minRadius for the lowest value to maxRadius for the highest value
points_values = 0:4
tracklist = BioCircosSNPTrack('mySNPTrack', points_chromosomes, points_coordinates,
points_values, colors = c("tomato2", "darkblue"), minRadius = 0.5, maxRadius = 0.9)
# Background are always placed below other tracks
tracklist = tracklist + BioCircosBackgroundTrack("myBackgroundTrack",
minRadius = 0.5, maxRadius = 0.9,
borderColors = "#AAAAAA", borderSize = 0.6, fillColors = "#B3E6FF")
tracklist = tracklist + BioCircosBackgroundTrack("myGenomeBackgroundTrack",
minRadius = 1.08, maxRadius = 1.1, fillColors = "#00FFFF")
BioCircos(tracklist, genomeFillColor = c("white", "white"),
chrPad = 0.05, displayGenomeBorder = FALSE, yChr = FALSE,
genomeTicksDisplay = FALSE, genomeLabelTextSize = 18, genomeLabelDy = 0)
Cheers.
@koalive thanks so much for this workaround