missing taxon_id column - bad upload from phyloseq
Hello,
I have been trying to upload and parse the data.
y <- parse_phyloseq(fil_nifH_out) Warning messages: 1: There is no "taxon_id" column in the data set "3", so there are no taxon IDs. 2: The data set "4" is named, but not named by taxon ids.
Then trying to filter out low counts
y$data$tax_data <- zero_low_counts(y, dataset = "tax_data", min_count = 5)
No cols specified and no numeric columns can be found.
Warning message:
No cols specified. No calculation will be done.
Any thoughts?
Does y have a dataset called "tax_data"? Can you show me the print out for y? Thanks
I think I know what is going on. In a phyloseq object "tax_data" is the taxonomic classifications. "otu_table" is the abundance matrix. If you dont say which columns you want to use, zero_low_counts looks for numeric columns. There are no numeric columns in y$data$tax_data, so it threw an error. Try:
y$data$otu_table <- zero_low_counts(y, dataset = "otu_table", min_count = 5)
That works no problem.
No cols specified, so using all numeric columns:
JZ030, JZ031, JZ032, JZ033, JZ034, JZ035, JZ036 ... JZ106, JZ108, JZ109, JZ110, JZ111, JZ112, JZ113
Zeroing 22423 of 287328 counts less than 5.
no_reads <- rowSums(y$data$tax_data[, otu_table$taxon_id]) == 0 sum(no_reads)
I am trying to make a heat tree based on site.
thank you for being so helpful. :-)
No problem! Note that the dataset option had its named changed to data in the new version. dataset will still work (for a while), but will give you a warning.
Sorry forgot to post the error no_reads <- rowSums(y$data$tax_data[, otu_table$taxon_id]) == 0 Error in otu_table$taxon_id : object of type 'closure' is not subsettable
sum(no_reads) Error: object 'no_reads' not found
y$data$tax_data[, otu_table$taxon_id] is the problem. In my example code, there is a sample data table with the names of samples. This was used to subset the abundance matrix to just the columns with sample counts.
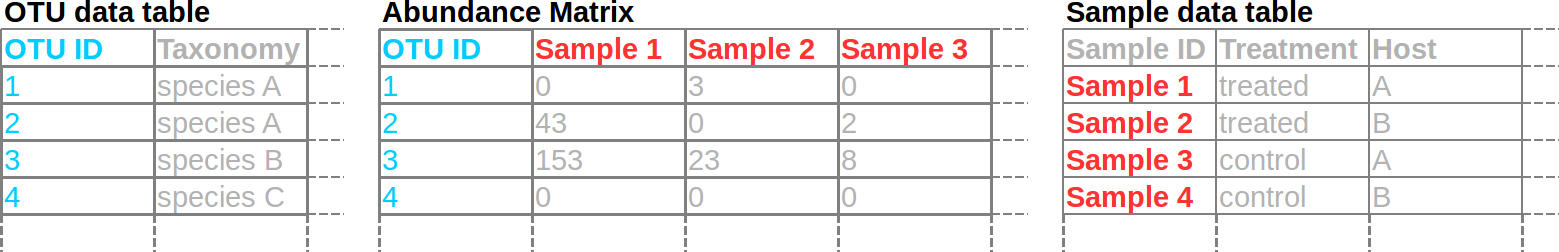
y$data$otu_table exists, but otu_table on its own is a phyloseq function (also known as a closure in R). You cant subset a function, so you get that error. You need to find a way to identify which columns in y$data$tax_data (should it be y$data$otu_data?) have count data so rowSums does not complain about non-numeric columns. A sample data table works well for this. Does y$data$sample_data have the column names for y$data$otu_data? Maybe something like:
no_reads <- rowSums(y$data$otu_data[, y$data$sample_data$sample_id]) == 0
Closing due to inactivity. If there are still unresolved issues, feel free to reopen this issue or open a new issue.
Hi Zachary,
Same problem and solved with your recommendation. Thank you. Maybe, you should correct it in the example:
https://grunwaldlab.github.io/metacoder_documentation/example.html
Hi @davidoctaviobotero,
What part should be corrected? There were a few problems discussed in the issue and I am not sure which you are talking about. Thanks
Hi Zachary,
I refer to change "tax_data" to "otu_table" in some lines in the example ( https://grunwaldlab.github.io/metacoder_documentation/example.html). Fro example, in the line of code for remove low-abundance counts:
no_reads <- rowSums(obj$data$tax_data[, hmp_samples$sample_id]) == 0sum(no_reads)
Should be:
no_reads <- rowSums(obj$data$otu_table[, hmp_samples$sample_id]) == 0sum(no_reads)
I didn't use these lines (I filtered and made these calcultaions using phyloseq) but I supposed has to be checked and correct also:
obj$data$tax_data <- zero_low_counts(obj, dataset = "tax_data", min_count = 5)
hmp_samples$inv_simp <- diversity(obj$data$tax_data[, hmp_samples$sample_id], index = "invsimpson", MARGIN = 2) # What orietation the matrix is in
Furthermore,
This one:
obj$data$tax_data <- calc_obs_props(obj, "tax_data")
Should be changed to:
obj$data$otu_table <- calc_obs_props(obj, "otu_table")
This one:
obj$data$tax_abund <- calc_taxon_abund(obj, "tax_data", cols = hmp_samples$sample_id)
Should be changed to:
obj$data$tax_abund <- calc_taxon_abund(obj, "otu_table", cols = hmp_samples$sample_id)
I hope these would be helpful,
David Octavio Botero Rozo, PhD
Cellular: +57 315 6490601
El vie., 16 de ago. de 2019 a la(s) 13:03, Zachary Foster ( [email protected]) escribió:
Hi @davidoctaviobotero https://github.com/davidoctaviobotero,
What part should be corrected? There were a few problems discussed in the issue and I am not sure which you are talking about. Thanks
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub https://github.com/grunwaldlab/metacoder/issues/248?email_source=notifications&email_token=AAQR5XNS3ELNFL4GKHW6L7LQE3TVLA5CNFSM4FSH7FA2YY3PNVWWK3TUL52HS4DFVREXG43VMVBW63LNMVXHJKTDN5WW2ZLOORPWSZGOD4PJWGA#issuecomment-522099480, or mute the thread https://github.com/notifications/unsubscribe-auth/AAQR5XP6FLNGESOEZ3IB54LQE3TVLANCNFSM4FSH7FAQ .
Ok, I see the issue now, thanks. The code is right in the sense that parse_tax_data returns a data set called tax_data by default, since it has no way of knowing that it is OTU data. When you start from phyloseq, the data set is called otu_data. Just think of it as a different variable name. It could be anything you want.
I will consider renaming the table to otu_data in the example so people know it can be changed.
I got it. Thank you!
David Octavio Botero Rozo, PhD
Cellular: +57 315 6490601
El mar., 20 de ago. de 2019 a la(s) 11:58, Zachary Foster ( [email protected]) escribió:
Ok, I see the issue now, thanks. The code is right in the sense that parse_tax_data returns a data set called tax_data by default, since it has no way of knowing that it is OTU data. When you start from phyloseq, the data set is called otu_data. Just think of it as a different variable name. It could be anything you want.
I will consider renaming the table to otu_data in the example so people know it can be changed.
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub https://github.com/grunwaldlab/metacoder/issues/248?email_source=notifications&email_token=AAQR5XM3IRHHRO5YNFKIDQ3QFQPDPA5CNFSM4FSH7FA2YY3PNVWWK3TUL52HS4DFVREXG43VMVBW63LNMVXHJKTDN5WW2ZLOORPWSZGOD4W633A#issuecomment-523103724, or mute the thread https://github.com/notifications/unsubscribe-auth/AAQR5XLETJVOVSRFKDWBSI3QFQPDPANCNFSM4FSH7FAQ .