Ask for a question about how to using AlphaFold Protein Structure Database to calculate the RSA?
Hi, Thanks for your great work! I‘m new to AlphaFold Protein Structure Database. Recently, I have performed a lysine labeling experiment and need to calculate the RSA (relative solvent accessibility) of lysine residues in human like the figure described below. There are only a few sentences which describe the RSA calculation. I have searched the database (https://alphafold.ebi.ac.uk/) but find no way to calculate RSA. It must be that I don't know how to use this database. Since I'm new here, I have no idea about how to perform this analysis. Here I sincerely ask for your help. Thanks a lot!
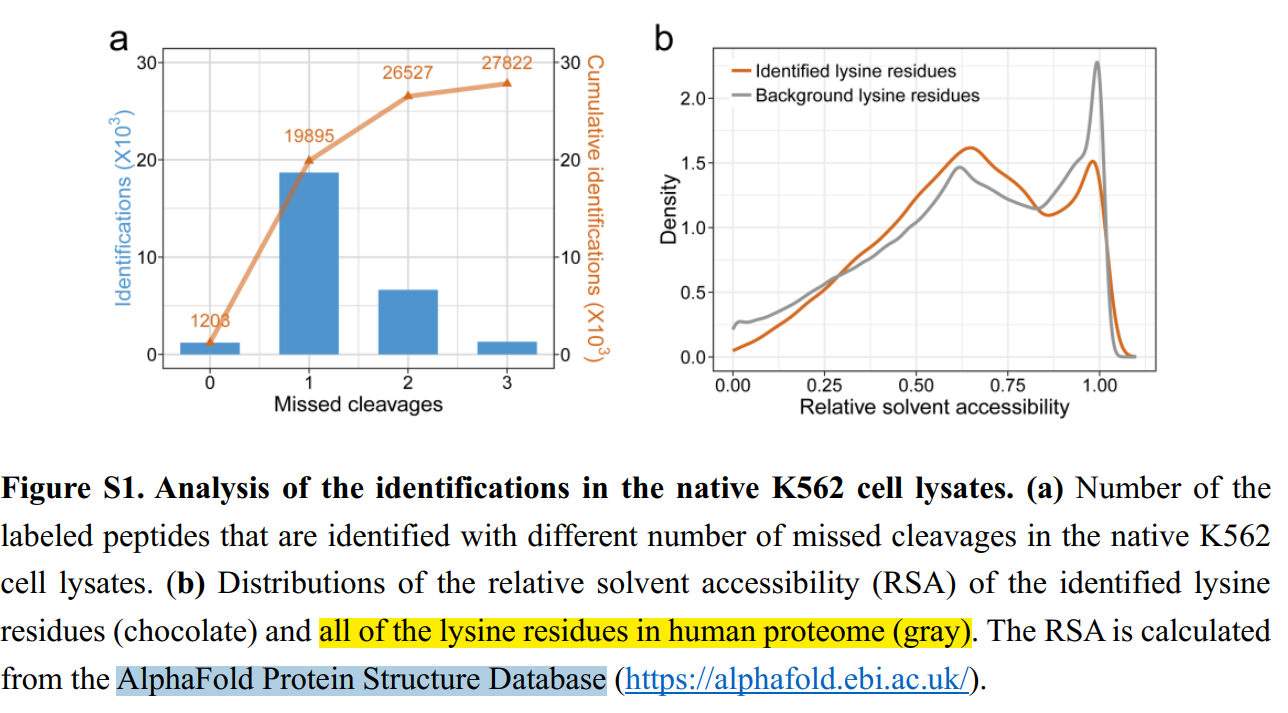
Pretty sure that they mean that the RSA was calculated on structures from the AlphaFold Protein Structure Database. However, there are many tools for calculating RSA, such as PyMOL, Rosetta (in all it's flavors), MOE, Discovery Studio, YASARA...