random prediction of resnet generated by hls4ml
Thank you for your great work! I am trying to generate Resnet20 with hls4ml. I trained Resnet20 with Qkeras and transfer to hls_model. when I ran hls_model.predict, it showed random prediction and the accuracy was low. But the quantized model works well. I tried with ap_fix<32,16> and ap_fix<16,6> but it still not work.
This is my model:
from tensorflow.keras import layers
from qkeras.qlayers import QDense, QActivation
from qkeras.quantizers import quantized_bits, quantized_relu
from qkeras import QActivation
from qkeras import QDense, QConv2DBatchnorm, QConv2D
from tensorflow.keras.layers import Dropout
subtract_pixel_mean = True
n = 3
version = 1
# Computed depth from supplied model parameter n
if version == 1:
depth = n * 6 + 2
elif version == 2:
depth = n * 9 + 2
# Model name, depth and version
model_type = 'ResNet%dv%d' % (depth, version)
def lr_schedule(epoch):
"""Learning Rate Schedule
Learning rate is scheduled to be reduced after 80, 120, 160, 180 epochs.
Called automatically every epoch as part of callbacks during training.
# Arguments
epoch (int): The number of epochs
# Returns
lr (float32): learning rate
"""
lr = 1e-3
if epoch > 180:
lr *= 0.5e-3
elif epoch > 160:
lr *= 1e-3
elif epoch > 120:
lr *= 1e-2
elif epoch > 80:
lr *= 1e-1
print('Learning rate: ', lr)
return lr
def resnet_layer(inputs,
num_filters=16,
kernel_size=3,
strides=1,
activation='relu',
batch_normalization=True,
conv_first=True, indx=0):
"""2D Convolution-Batch Normalization-Activation stack builder
# Arguments
inputs (tensor): input tensor from input image or previous layer
num_filters (int): Conv2D number of filters
kernel_size (int): Conv2D square kernel dimensions
strides (int): Conv2D square stride dimensions
activation (string): activation name
batch_normalization (bool): whether to include batch normalization
conv_first (bool): conv-bn-activation (True) or
bn-activation-conv (False)
# Returns
x (tensor): tensor as input to the next layer
"""
x = inputs
if conv_first:
if batch_normalization:
x = QConv2DBatchnorm(num_filters,
kernel_size=kernel_size,
kernel_quantizer="quantized_bits(8,0,alpha=1)",
bias_quantizer="quantized_bits(8,0,alpha=1)",
strides=strides,
padding='same',
kernel_initializer='he_normal',
kernel_regularizer=l2(1e-4), name= 'fused_convbn{}_1'.format(indx))(x)
else:
x = QConv2D(num_filters,
kernel_size=kernel_size,
kernel_quantizer="quantized_bits(8,0,alpha=1)",
bias_quantizer="quantized_bits(8,0,alpha=1)",
strides=strides,
padding='same',
kernel_initializer='he_normal',
kernel_regularizer=l2(1e-4), name= 'conv{}_1'.format(indx))(x)
if activation is not None:
x = QActivation('quantized_relu(8)')(x)
else:
if batch_normalization:
x = BatchNormalization()(x)
if activation is not None:
x = QActivation('quantized_relu(8)')(x)
x = QConv2D(num_filters,
kernel_size=kernel_size,
kernel_quantizer="quantized_bits(8,0,alpha=1)",
bias_quantizer="quantized_bits(8,0,alpha=1)",
strides=strides,
padding='same',
kernel_initializer='he_normal',
kernel_regularizer=l2(1e-4), name= 'conv{}_2'.format(indx))(x)
return x
def resnet_v1(input_shape, depth, num_classes=10):
if (depth - 2) % 6 != 0:
raise ValueError('depth should be 6n+2 (eg 20, 32, 44 in [a])')
# Start model definition.
num_filters = 16
num_res_blocks = int((depth - 2) / 6)
inputs = Input(shape=input_shape)
indx = 0
x = resnet_layer(inputs=inputs)
indx = indx +1
# Instantiate the stack of residual units
for stack in range(3):
for res_block in range(num_res_blocks):
strides = 1
if stack > 0 and res_block == 0: # first layer but not first stack
strides = 2 # downsample
y = resnet_layer(inputs=x,
num_filters=num_filters,
strides=strides,indx=indx)
indx = indx + 1
y = resnet_layer(inputs=y,
num_filters=num_filters,
activation=None,indx=indx)
indx = indx + 1
if stack > 0 and res_block == 0: # first layer but not first stack
# linear projection residual shortcut connection to match
# changed dims
x = resnet_layer(inputs=x,
num_filters=num_filters,
kernel_size=1,
strides=strides,
activation=None,
batch_normalization=False, indx=indx)
indx = indx +1
x = keras.layers.add([x, y])
x = QActivation('quantized_relu(8)')(x)
num_filters *= 2
# Add classifier on top.
# v1 does not use BN after last shortcut connection-ReLU
x = AveragePooling2D(pool_size=8)(x)
# x = MaxPooling2D(pool_size=8)(x)
y = Flatten(name = 'flattenlayer')(x)
x = Dense(num_classes,
activation=None,
kernel_initializer='he_normal', name ='output_dense')(y)
outputs = Activation('softmax',name='output_softmax')(x)
# Instantiate model.
model = Model(inputs=inputs, outputs=outputs)
return model
model = resnet_v1(input_shape=input_shape, depth=depth)
model.compile(loss='categorical_crossentropy',
optimizer=Adam(lr=lr_schedule(0)),
metrics=['accuracy'])
This is how I train the model:
# Prepare model model saving directory.
save_dir = os.path.join(os.getcwd(), 'saved_models')
model_name = 'cifar10_%s_model.{epoch:03d}.h5' % model_type
if not os.path.isdir(save_dir):
os.makedirs(save_dir)
filepath = os.path.join(save_dir, model_name)
# Prepare callbacks for model saving and for learning rate adjustment.
checkpoint = ModelCheckpoint(filepath=filepath,
monitor='val_acc',
verbose=1,
save_best_only=True)
lr_scheduler = LearningRateScheduler(lr_schedule)
lr_reducer = ReduceLROnPlateau(factor=np.sqrt(0.1),
cooldown=0,
patience=5,
min_lr=0.5e-6)
callbacks = [checkpoint, lr_reducer, lr_scheduler]
# Run training, with or without data augmentation.
if not data_augmentation:
print('Not using data augmentation.')
model.fit(x_train, y_train,
batch_size=batch_size,
epochs=epochs,
validation_data=(x_test, y_test),
shuffle=True,
callbacks=callbacks)
else:
print('Using real-time data augmentation.')
# This will do preprocessing and realtime data augmentation:
datagen = ImageDataGenerator(
# set input mean to 0 over the dataset
featurewise_center=False,
# set each sample mean to 0
samplewise_center=False,
# divide inputs by std of dataset
featurewise_std_normalization=False,
# divide each input by its std
samplewise_std_normalization=False,
# apply ZCA whitening
zca_whitening=False,
# epsilon for ZCA whitening
zca_epsilon=1e-06,
# randomly rotate images in the range (deg 0 to 180)
rotation_range=0,
# randomly shift images horizontally
width_shift_range=0.1,
# randomly shift images vertically
height_shift_range=0.1,
# set range for random shear
shear_range=0.,
# set range for random zoom
zoom_range=0.,
# set range for random channel shifts
channel_shift_range=0.,
# set mode for filling points outside the input boundaries
fill_mode='nearest',
# value used for fill_mode = "constant"
cval=0.,
# randomly flip images
horizontal_flip=True,
# randomly flip images
vertical_flip=False,
# set rescaling factor (applied before any other transformation)
rescale=None,
# set function that will be applied on each input
preprocessing_function=None,
# image data format, either "channels_first" or "channels_last"
data_format=None,
# fraction of images reserved for validation (strictly between 0 and 1)
validation_split=0.0)
# Compute quantities required for featurewise normalization
# (std, mean, and principal components if ZCA whitening is applied).
datagen.fit(x_train)
# Fit the model on the batches generated by datagen.flow().
model.fit_generator(datagen.flow(x_train, y_train, batch_size=batch_size),
validation_data=(x_test, y_test),
epochs=50, verbose=1, workers=4,
callbacks=callbacks)
# Score trained model.
scores = model.evaluate(x_test, y_test, verbose=1)
print('Test loss:', scores[0])
print('Test accuracy:', scores[1])
**This is the config:**
hls_config = hls4ml.utils.config_from_keras_model(model, granularity='name')
hls_config['Model']['Strategy'] = 'Resource'
hls_config['Model']['ReuseFactor'] = 2
hls_config['Model']['Precision'] = 'ap_fixed<16,6>'
for Layer in hls_config['LayerName'].keys():
if Layer == "flattenlayer":
hls_config['LayerName'][Layer]['Strategy'] = 'Resource'
hls_config['LayerName'][Layer]['Precision'] = 'ap_fixed<32,16>'
if Layer == "output_dense":
hls_config['LayerName'][Layer]['Strategy'] = 'Resource'
hls_config['LayerName'][Layer]['Precision'] = 'ap_fixed<32,16>'
if Layer == "output_softmax":
hls_config['LayerName'][Layer]['Strategy'] = 'Stable'
hls_config['LayerName'][Layer]['Precision'] = 'ap_fixed<32,16>'
hls_model = hls4ml.converters.convert_from_keras_model(model,
backend='VivadoAccelerator',
io_type='io_stream',
hls_config=hls_config,
output_dir='model_1/hls4ml_prj',
board = 'zcu102',
part='xczu9eg-ffvb1156-2-e'
) #xcu280-fsvh2892-2L-e
I test like this:
hls_model.compile()
print("---------compile finish--------------------------")
from sklearn.metrics import accuracy_score
def plotROC(Y, y_pred, y_pred_hls4ml, label="Model"):
accuracy_keras = float(accuracy_score (np.argmax(Y,axis=1), np.argmax(y_pred,axis=1)))
accuracy_hls4ml = float(accuracy_score (np.argmax(Y,axis=1), np.argmax(y_pred_hls4ml,axis=1)))
print("Accuracy Keras: {}".format(accuracy_keras))
print("Accuracy hls4ml: {}".format(accuracy_hls4ml))
fig, ax = plt.subplots(figsize=(9, 9))
# _ = plotting.makeRoc(Y, y_pred, labels=['%i'%nr for nr in range(n_classes)])
plt.gca().set_prop_cycle(None) # reset the colors
_ = plotting.makeRoc(Y, y_pred_hls4ml, labels=['%i'%nr for nr in range(10)], linestyle='--')
from matplotlib.lines import Line2D
lines = [Line2D([0], [0], ls='-'),
Line2D([0], [0], ls='--')]
from matplotlib.legend import Legend
leg = Legend(ax, lines, labels=['Keras', 'hls4ml'],
loc='lower right', frameon=False)
ax.add_artist(leg)
plt.figtext(0.2, 0.38,label, wrap=True, horizontalalignment='left',verticalalignment='center')
plt.ylim(0.01,1.)
plt.xlim(0.7,1.)
X_test_reduced = x_test[:50]
Y_test_reduced = y_test[:50]
start = time.time()
y_predict = model.predict(X_test_reduced)
end = time.time()
print('\n model took {} minutes to run!\n'.format( (end - start)/60.))
start = time.time()
y_predict_hls4ml = hls_model.predict(np.ascontiguousarray(X_test_reduced))
end = time.time()
print('\n hls model took {} minutes to run!\n'.format( (end - start)/60.))
plotROC(Y_test_reduced, y_predict, y_predict_hls4ml, label="Keras")
resnet20_1-QKeras-Copy2.ipynb.zip
Attached is my code. I will be grateful if you could pls help me with it.
I think there might be some problem with the last softmax layer. I printed the output of "output_softmax" and it showed like the fig.
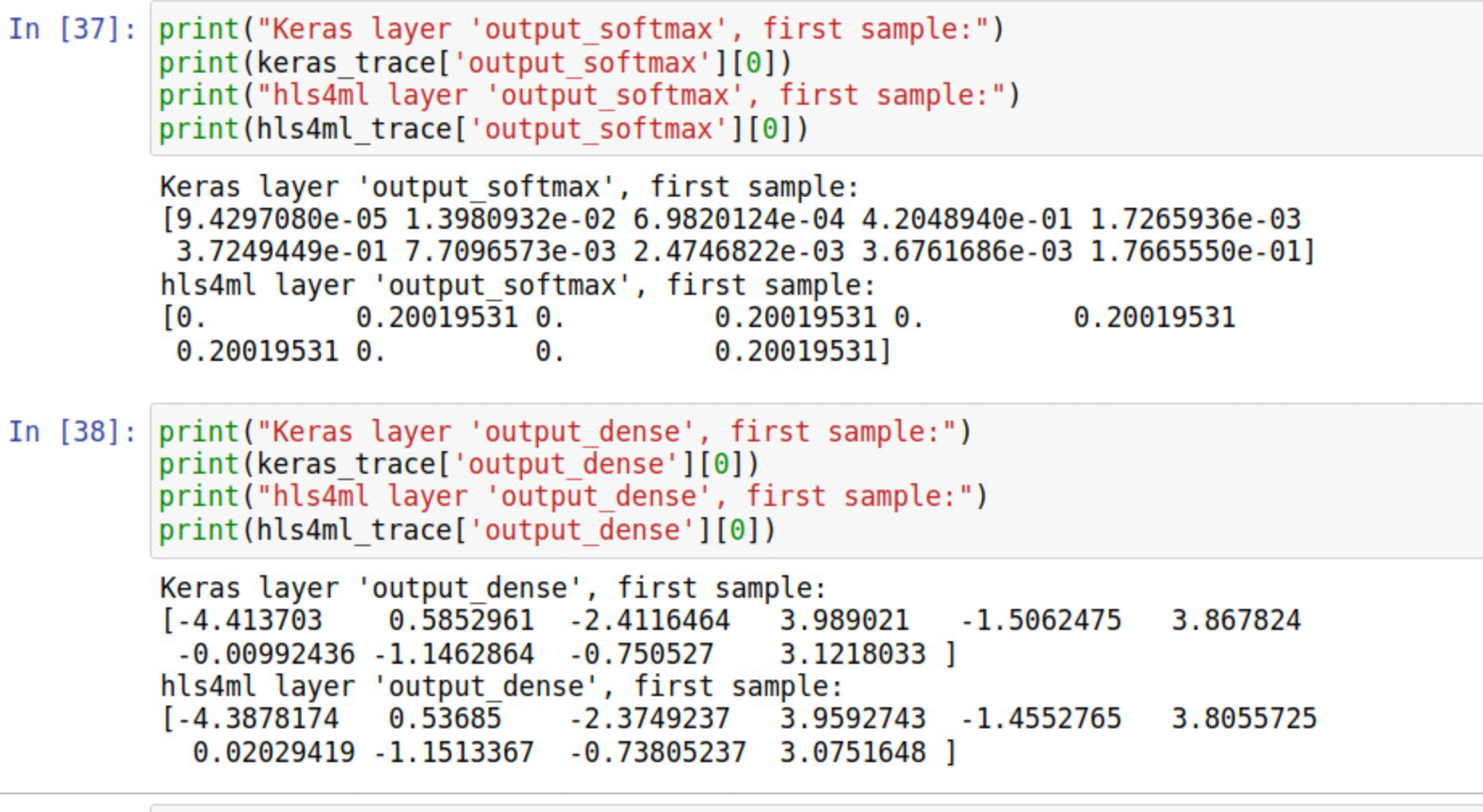
Similar issues happened on my side, What is the current solution or workaround? @liuhao-97