LDhat
 LDhat copied to clipboard
LDhat copied to clipboard
Estimate recombination rates from population genetic data
Dear developers, [errlog.txt](https://github.com/auton1/LDhat/files/13933940/errlog.txt) I am not sure if this program is still being maintained but it might be good to enforce `-std=c++11` in the make file, e.g. `CFLAGS = -O3...
Hello, We have used vcftools to convert vcf's that were generated from a simulation using SLiM (simulated genome with only As and Ts) to ldhat format, and we are getting...
Thank you for your software.I'm trying to use software to calculate the hotpots of recombination in the MHC region. THE SNP data of chr6 MHC region of YRI population in...
Hi there I am working on a diploid grass population and trying to estimate recombination rate by "interval" program. Since this population has three major genetic clusters, therefore, I tried...
Hello. I plan to use LDhat to estimate recombination rate along the chromosomes wild wheat which is a selfer species. The data is composed of ~400 genotypes sampled from ~100...
Hello, I ran _interval_ on 2000 SNPs of a region about 300 Kb. I have 10 diploid individuals. I ran interval with block penalty of 5, 10 million iterations and...
Hello, I was wondering how I could use the `interval` program of LDhat to generate sex-averaged rho for sex chromosomes where females are diploid and males are haploid? Having 5...
Hi, I am working on a non model organism and I am looking to estimate recombination rate along my genome and different part of the genome (ex chromosome, within chromosome...
Hello, Thank you for your software. I was wondering what does it mean when the composite likelihood plateaus for different values of rho as shown in the figure: 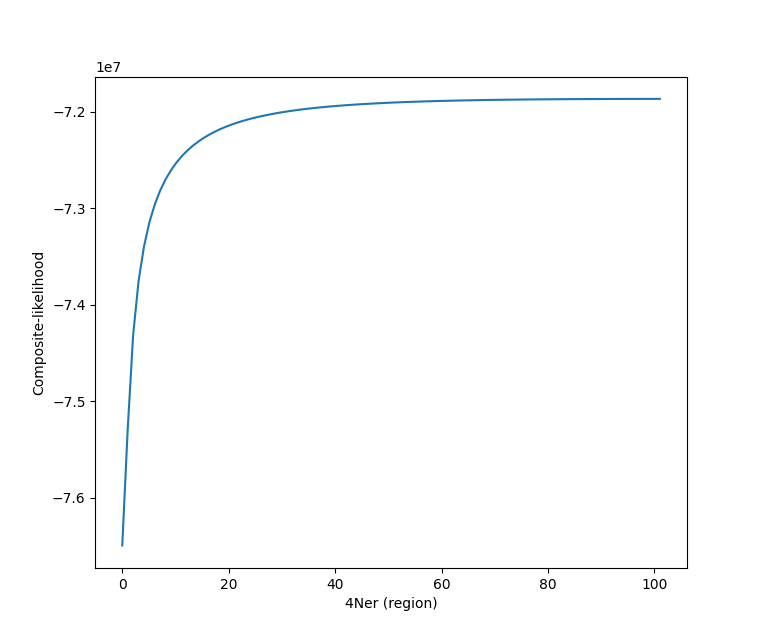 Thank...
In case anyone is interested, I created a new likelihood lookup table for n = 320 sequences/chromosomes -- see https://zenodo.org/record/3934350 That seemed sufficiently large and computationally expensive to make sharing...
