EnrichKegg still shows all pvalues even after specifying a specific pvalue cutoff
Although I use pvalueCutoff, the results still show pvalues larger than 0.1. Why is that?
kk2 <- enrichKEGG(gene = dedup_ids_gene$ENTREZID, universe=dedup_ids_bg$ENTREZID, keyType = "ncbi-geneid", organism = kegg_organism, pvalueCutoff = 0.1, pAdjustMethod = "fdr", use_internal_data = FALSE)
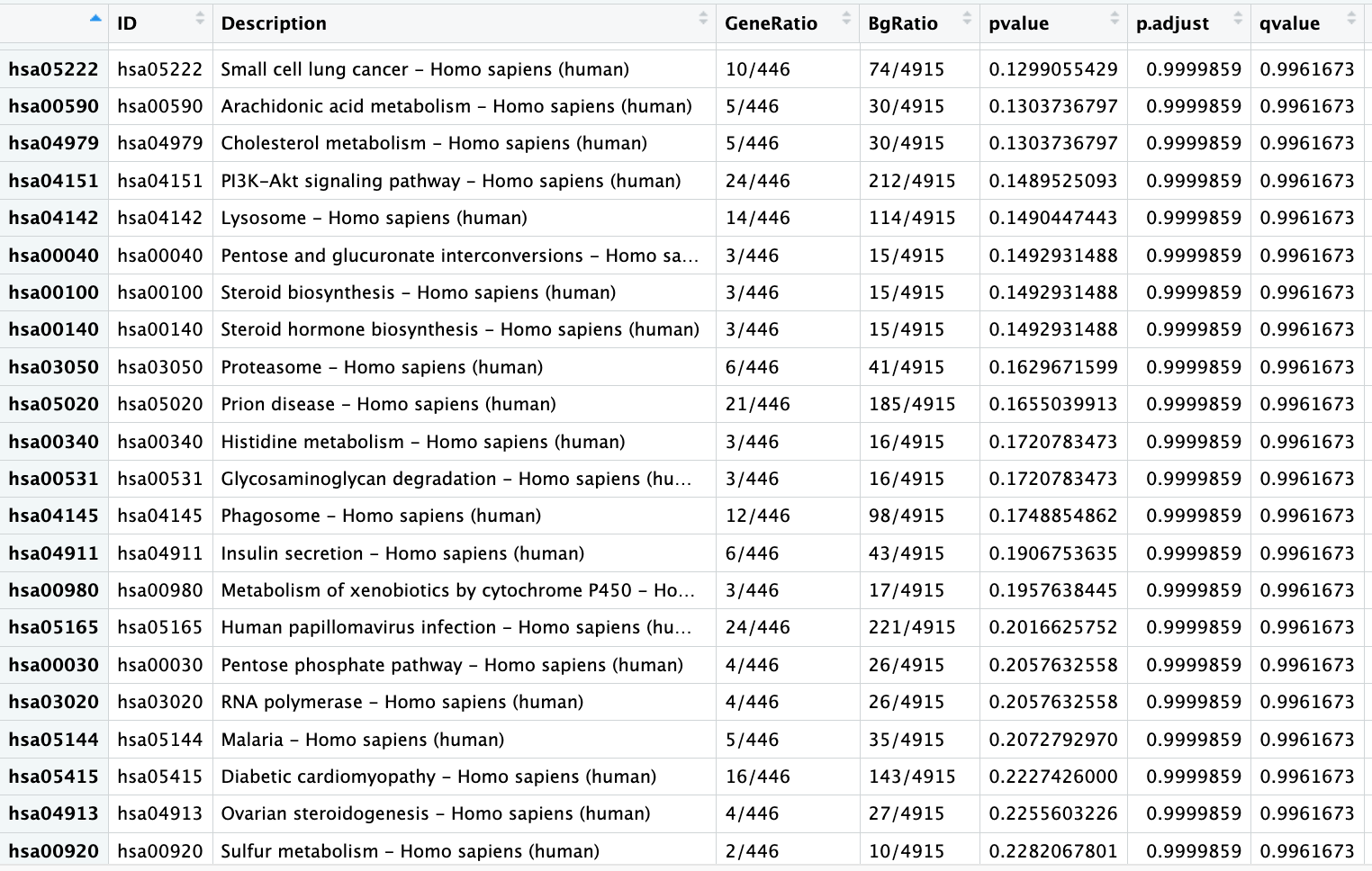
I am experiencing this same issue for all the enrichment functions. I am currently just filtering the p.adjust column manually, but surely this is not intended?
Please provide the kk2 object, and see the result of as.data.frame(kk2).
Hello, the as.data.frame() call works to appropriately filter the data matrix for me. To me, this was not clear from the current documentation. Thank you for the explanation.