Error when emapplot the selected results of clusterprofiler
When I emapplot the selected results of clusterprofiler, I got the "Error in if (n == 1) { : the condition has length > 1". Can you help me to resolve it ? There are my code:
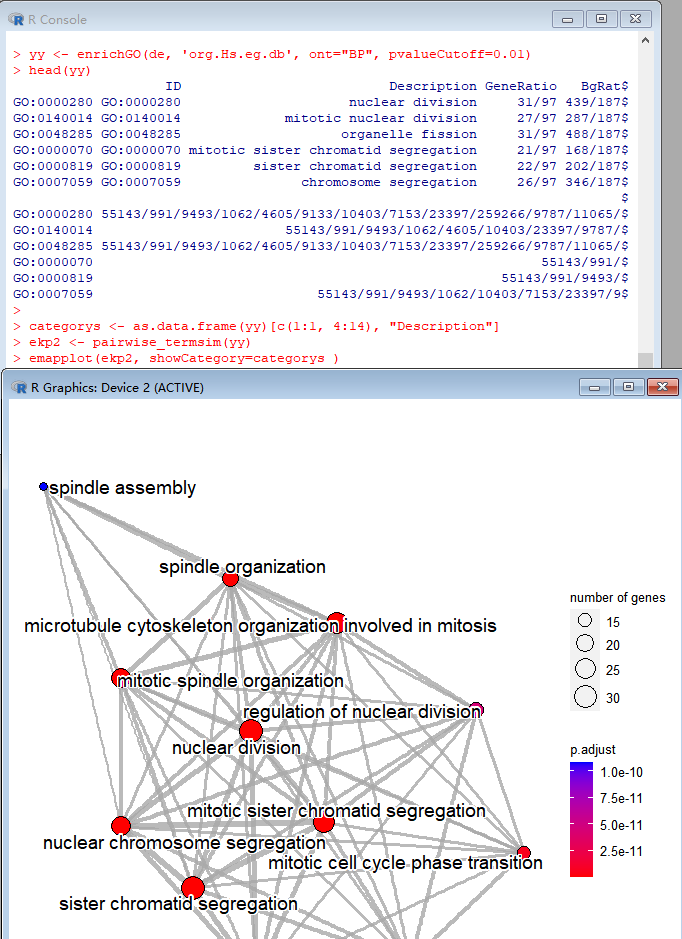
categorys <- as.data.frame(ekp)[c(1:1, 4:14), "Description"] ekp2 <- pairwise_termsim(ekp) emapplot(ekp2, showCategory=categorys )
Please provide repeatable examples(with your data) and use the latest version of enrichplot.
Please provide repeatable examples(with your data) and use the latest version of
enrichplot.Thanks for your reply. I used enricher to perform kegg enrichment analysis. dotplot and cnetplot can both be performed with "showCategory=categorys", only emapplot reported "Error in if (n == 1) { : the condition has length > 1".There are my all code:
pathway2gene <- read.table("./pathway2gene",header = T,sep = "\t") pathway2name <- read.table("./pathway2name",header = T,sep = "\t") gene <- read.csv("./DEP_liver_FC0.58_up.csv") gene_list <- gene[,2] ekp <- enricher(gene_list, TERM2GENE = pathway2gene, TERM2NAME = pathway2name, pvalueCutoff = 0.05, qvalueCutoff = 0.05, pAdjustMethod = "BH", minGSSize = 1) categorys <- as.data.frame(ekp)[c(1:1, 4:14), "Description"] dotplot(ekp,color="p.adjust", showCategory = categorys, font.size=10, title = 'KEGG Pathway') cnetplot(ekp,showCategory=categorys) ekp2 <- pairwise_termsim(ekp) library(ggnewscale) emapplot(ekp2, showCategory = categorys )
I run the example code of emapplot, the same error was also reported. Is this a bug?
Example:
library(DOSE)
data(geneList)
de <- names(geneList)[1:100]
x <- enrichDO(de)
x2 <- pairwise_termsim(x)
emapplot(x2)
# use layout to change the layout of map
emapplot(x2, layout = "star")
# use showCategory to select the displayed terms. It can be a number of a vector of terms.
emapplot(x2, showCategory = 10)
categorys <- c("pre-malignant neoplasm", "intestinal disease",
"breast ductal carcinoma", "non-small cell lung carcinoma")
emapplot(x2, showCategory = categorys)

@ppdpg

categorys <- intersect(categorys, x2$Description)
categorys
emapplot(x2, showCategory = categorys)
It is a typo in the example: should be "lung non-small cell carcinoma", not "non-small cell lung carcinoma". We will fix the document in the next version. Thanks.
Thanks for your reply. However, after performed "categorys <- intersect(categorys, x2$Description)", the "Error in if (n == 1) { : the condition has length > 1" still reported when I performed "emapplot(x2, showCategory = categorys)".
I don't know why this mistake occurred.
I am also having this issue
It will be fixed in the next version of enrichplot.