ENH: Add Annotation Store Reader
This PR adds an AnnotationStoreReader class that allows an annotation store to be read as if it were a WSI. That can be used wherever a WSIreader can be used.
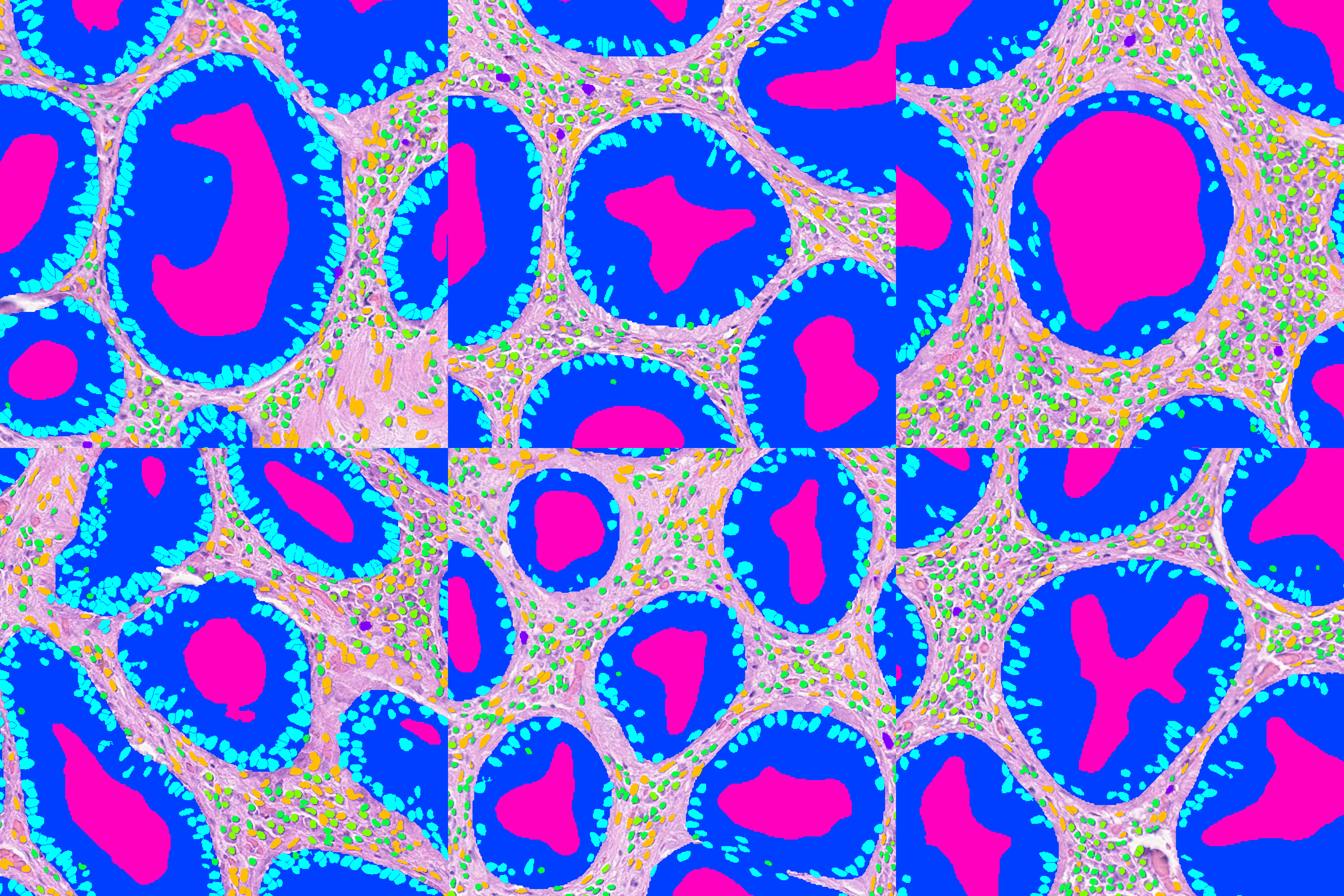
For example, to extract overlay patches around a few random glands using PointsPatchExtractor:
SQ = SQLiteStore(path\to\store.db)
glands = SQ.query(where='props["type"] == "Gland"')
centroids = np.array([[gland.geometry.centroid.x, gland.geometry.centroid.y] for gland in glands.values()])
#get 6 random centroids
rand_centroids = centroids[np.random.randint(0, len(centroids), 6),:]
ann_reader = AnnotationStoreReader(db_path, base_wsi_reader=wsi)
extractor = PointsPatchExtractor(ann_reader, rand_centroids, patch_size=(1024, 1024))
#show the 6 patches in a 2x3 grid
patch_im = np.zeros((2048, 3072, 3), dtype=np.uint8)
for i, patch in enumerate(extractor):
patch_im[1024*(i//3):1024*(i//3 + 1), (i%3)*1024:(i%3+1)*1024, :] = patch
plt.imshow(patch_im)
plt.show()
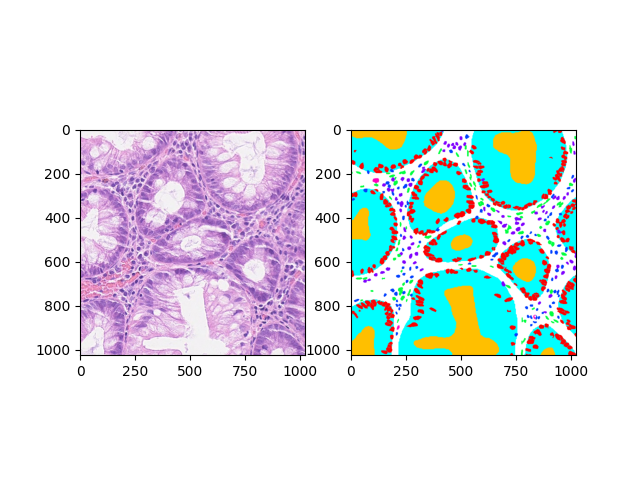
Can be used to draw annotations over its base wsi as an 'annotated slide' as above, or just as a standalone overlay mask as below right.
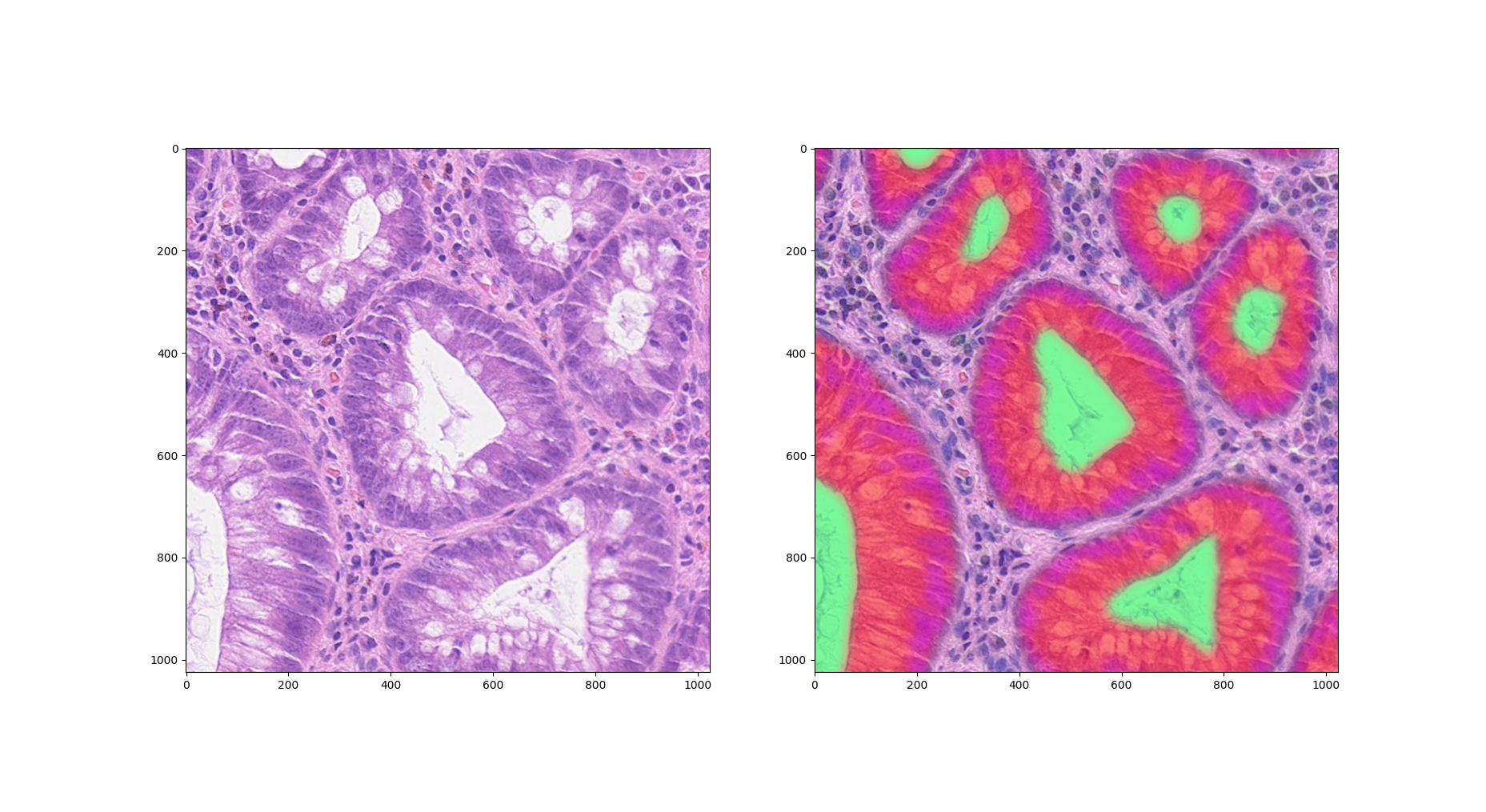
Also moves the annotation rendering code into the AnnotationRender class instead of in the AnnotationTileGenerator, so it can be reused as needed. It probably belongs more naturally there regardless.
Updates the rendering code a little as well with a few further options, for example options for a blur to give a more heatmap-like effect:
Codecov Report
Merging #476 (d8af126) into develop (1f0c8ea) will increase coverage by
0.03%. The diff coverage is100.00%.
@@ Coverage Diff @@
## develop #476 +/- ##
===========================================
+ Coverage 99.63% 99.67% +0.03%
===========================================
Files 62 62
Lines 6378 6487 +109
Branches 1019 1074 +55
===========================================
+ Hits 6355 6466 +111
+ Misses 10 9 -1
+ Partials 13 12 -1
| Impacted Files | Coverage Δ | |
|---|---|---|
| tiatoolbox/wsicore/wsimeta.py | 100.00% <ø> (ø) |
|
| tiatoolbox/annotation/storage.py | 99.87% <100.00%> (ø) |
|
| tiatoolbox/tools/pyramid.py | 100.00% <100.00%> (ø) |
|
| tiatoolbox/utils/visualization.py | 100.00% <100.00%> (ø) |
|
| tiatoolbox/wsicore/wsireader.py | 98.47% <100.00%> (+0.39%) |
:arrow_up: |
| tiatoolbox/utils/image.py | 100.00% <0.00%> (ø) |
:mega: We’re building smart automated test selection to slash your CI/CD build times. Learn more
Please see comments on other PR:
- https://github.com/TissueImageAnalytics/tiatoolbox/pull/445#issuecomment-1271385492
- https://github.com/TissueImageAnalytics/tiatoolbox/pull/445#issuecomment-1271392213