Predicted labels do not appear.
Hello, Thank you for your work again. I would like to know how the test data annotated. I have annotated Kitti raw data using https://github.com/jbehley/point_labeler this tool and successfully trained my model. However, after testing the raw sequences and getting the labels when I open the predicted label in annotation tool, it does not give any prediction.

Just looks like everything is gray.
I have tried to change def to_label in confidences_to_label.py. But i have no luck. Can you please look into this ? Thank you.
Hi @E4CEeg,
Did you verify that your annotated data is correct? One way to check would be to use the visualizer of the SemanticKITTI API. You can also use this tool to visualize your MOS predictions. I never used the point labeler to visualize predictions.
Best Benedikt
Did you find any problem on our side? Otherwise, I would close this issue since it seems to be related to the point labeling rather than the 4DMOS code.
Hello,
As you have suggested, I have tried visualizer of the SemanticKitti API. My annotated labels are shown in the visualizer. However, my predictions are not shown it. First, I have trained the model with my annotated data. With my trained model, I have tried to predict my own custom data and sequence 11 of SemanticKitti. Both of them do not show any result in the visualizer. Then I have used 10scans.ckpt to predict my own data and same as sequence 11 from SemanticKitti. However, still the results of both of them do not show any moving object.
Which labels are you using for annotation?
It should definitely work predicting moving objects with the pre-trained 10scans.ckpt model on sequence 11 and visualizing them with the SemanticKITTI API. Can you post the commands you use for running the inference scripts and what your predictions folder looks like?
Hello, sorry for the late reply.
I am also confused about the result. So I have started again, using only the pre-trained model 10scans and semantickitti sequence 11.
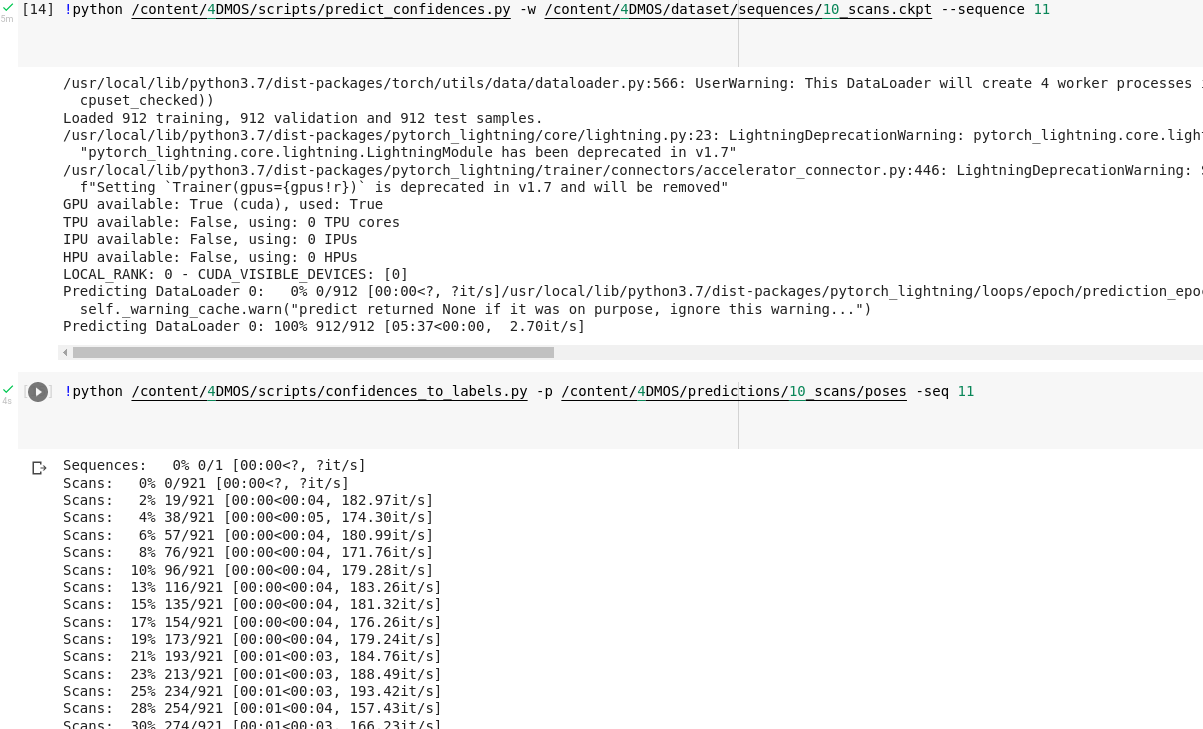
Here i am running the given trained model.
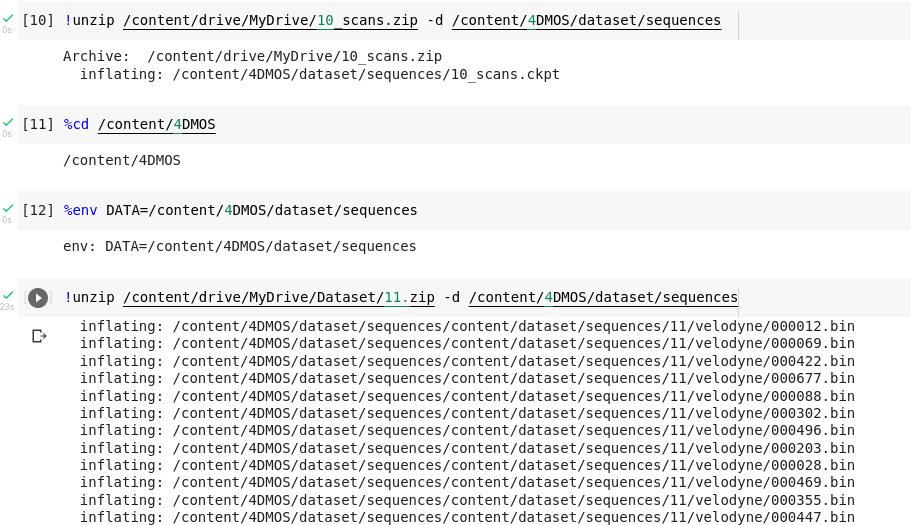
 Here is the folder file structure.
Here is the folder file structure.
 Here , I am running the visualizer.
Here , I am running the visualizer.
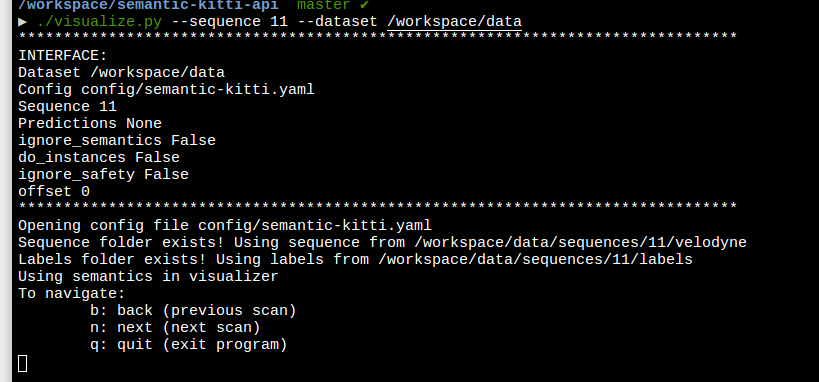 And this is the shown result. There is a moving van in between scan21-31.
This is the scan 22.
And this is the shown result. There is a moving van in between scan21-31.
This is the scan 22.
 This is the scan 29
This is the scan 29

Unfortunately, in the visualizer, nothing shows up.
By running visualize.py, you are using the semantic segmentation config of SemanticKITTI and not the MOS config file. The problem is the wrong label mapping since moving objects are labeled with the class ID 251 which is not present in the SemanticKITTI config for semantic segmentation. Therefore, no labels are displayed. You need to run either
./visualize_mos.py --sequence 8 --dataset /path/to/dataset
while providing labels in the labels folder or you specify the path to the predictions as described in the README:
./visualize_mos.py --sequence 8 --dataset /path/to/dataset --predictions /path/to/4DMOS/predictions/ID/POSES/labels/STRATEGY/
Oh you're right. That was a very stupid error. Thank you very much.