Length filtering still performed even if --disable_length_filtering is turned on
I wasn't expecting any filtering to happen if I specify these options --disable_trim_poly_g --disable_length_filtering --disable_quality_filtering. However, the HTML always reports that some small percentage of reads is filtered due to being too short. Is this intended or...?
Hi @uros-sipetic
That's probably because those reads being filtered with failed_too_short even with filtering options disabled, were actually fully trimmed from the 5' end, due to low base quality at the first four bases.
I used to get confused by this, too. However, when I specified the --failed_out option and look into the failed reads with the failure reason failed_too_short, I noticed that all that kind of reads have very low base quality at the first four reads on 5' end, as below:
@SRR10260015.759 D00562:262:C9G0HANXX:4:2213:8841:2053/2 failed_too_short
TTGAAGAGAATTTTTGTTGGGGTTTTGTGAAAATATTTTATATTTAATAAAAAAAAAAAATAAAAATCCTAGGGGATTGTTAAATCAACCCCCCTCCCTCTCTTATTTTTTTTTTTATATTTTAT
+
333001111111111/11;0/////011111:1:1<111111111111?<@1CEG<////00=:000000000<///00><0000;0000..8..:.:...9/:////66C..C.8.///6////
Unfortunately, if you also choose to perform the sliding window cutting by evaluating the mean quality scores in the sliding window, specifically by using -r, --cut_right, the whole sequence will be dropped from its 5' end, as the default sliding window size is four, and the default mean quality value for each window is 20.
Just it could be confusing to label the same failure reason for reads that are too short after trimming (more than 1 nt), as well as reads that are fully discarded after trimming. Also default filtering of the latter kind of reads is not reported in the stderr as 'reads failed due to too short: xxx', which raises inconsistency between the txt summary and html report, as well as confusions like 'how could I miss those reads since no filterings have been performed?'.
Thus, @sfchen, I recommend some improvements on this. Either more detailed description of this potential length filtering in readme.md, or in the report could be very helpful.
Hi, I'm having a similar issue for paired end fastp trimming when I'm passing --adapter_fasta. Reads are discarded as too short even if I attempt to disable length filtering by --disable_length_filtering or --length_required 0.
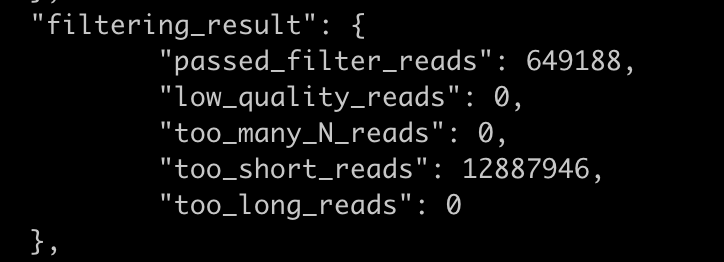
Please let me know how to properly override this filtering step. Thank you , Geoff
I meet the same problem, seems even disable all fastp provided filter option, there still many reads failed_too_short (looks like length 0) in failed output.... @sfchen any idea it? please let me know, thanks...