SVM-ANN
 SVM-ANN copied to clipboard
SVM-ANN copied to clipboard
The code for NIRS analysis based SVM&ANN
SVM-ANN
The code for NIRS analysis based SVM&ANN
一、数据来源
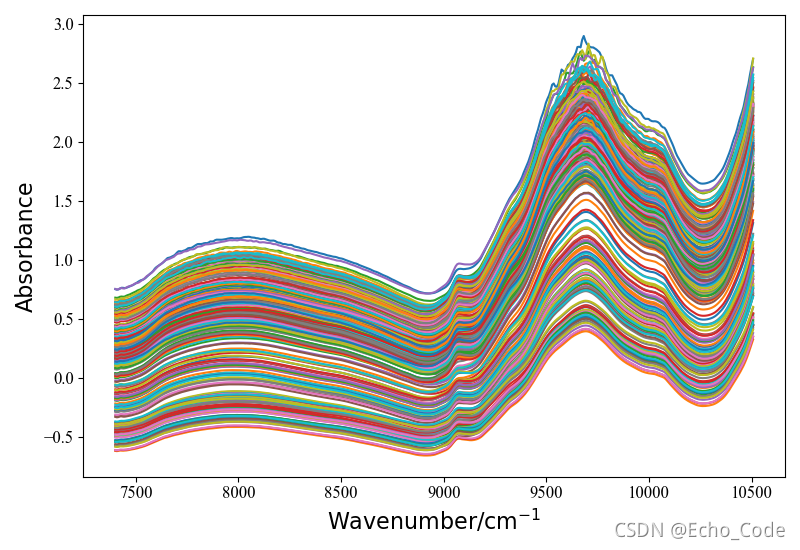
使用药品数据,共310个样本,每条样本404个变量,根据活性成分,分成4类
图片如下:

:
# figsize = 5, 3
figsize = 8, 5.5
figure, ax = plt.subplots(figsize=figsize)
# ax = plt.figure(figsize=(5,3))
x_col = x_col[::-1] # 数组逆序
y_col = np.transpose(data_x)
plt.plot(x_col, y_col )
plt.tick_params(labelsize=12)
labels = ax.get_xticklabels() + ax.get_yticklabels()
[label.set_fontname('Times New Roman') for label in labels]
font = {'weight': 'normal',
'size': 16,
}
plt.xlabel("Wavenumber/$\mathregular{cm^{-1}}$", font)
plt.ylabel("Absorbance", font)
# plt.title("The spectrum of the {} dataset".format(tp), fontweight="semibold", fontsize='x-large')
plt.show()
plt.tick_params(labelsize=23)
def TableDataLoad(tp, test_ratio, start, end, seed):
# global data_x
data_path = '..//Data//table.csv'
Rawdata = np.loadtxt(open(data_path, 'rb'), dtype=np.float64, delimiter=',', skiprows=0)
table_random_state = seed
if tp =='raw':
data_x = Rawdata[0:, start:end]
# x_col = np.linspace(0, 400, 400)
if tp =='SG':
SGdata_path = './/Code//TableSG.csv'
data = np.loadtxt(open(SGdata_path, 'rb'), dtype=np.float64, delimiter=',', skiprows=0)
data_x = data[0:, start:end]
if tp =='SNV':
SNVata_path = './/Code//TableSNV.csv'
data = np.loadtxt(open(SNVata_path, 'rb'), dtype=np.float64, delimiter=',', skiprows=0)
data_x = data[0:, start:end]
if tp == 'MSC':
MSCdata_path = './/Code//TableMSC.csv'
data = np.loadtxt(open(MSCdata_path, 'rb'), dtype=np.float64, delimiter=',', skiprows=0)
data_x = data[0:, start:end]
data_y = Rawdata[0:, -1]
x_col = np.linspace(7400, 10507, 404)
plotspc(x_col, data_x[:, :], tp=0)
x_data = np.array(data_x)
y_data = np.array(data_y)
X_train, X_test, y_train, y_test = train_test_split(x_data, y_data, test_size=test_ratio,random_state=table_random_state)
return X_train, X_test, y_train, y_test
2.1 基于SVM的药品光谱分类
2.1.1 建立SVM参数寻找,找到最佳的SVM参数
def SVM_Classs_test(train_x ,train_y,test_x,test_y):
params = [
{'kernel': ['linear'], 'C': [ 0.1,0.5, 1, 1.5,2,3,5, 10, 15,50,100],'gamma': [1e-7,1e-6,1e-5, 1e-4, 1e-3, 1e-2, 1e-1, 1]},
{'kernel': ['poly'], 'C': [0.1, 1, 2, 10, 15], 'degree': [2, 3]}
# {'kernel': ['rbf'], 'C': [0.1, 1, 2, 10], 'gamma':[1e-3, 1e-2, 1e-1, 1, 2]}
]
model = ms.GridSearchCV(svm.SVC(probability=True),
params,
refit=True,
return_train_score=True, # 后续版本需要指定True才有score方法
cv=10)
model.fit(train_x, train_y)
model_best = model.best_estimator_
pred_test_y = model_best.predict(test_x)
acc_test = accuracy_score(test_y, pred_test_y)
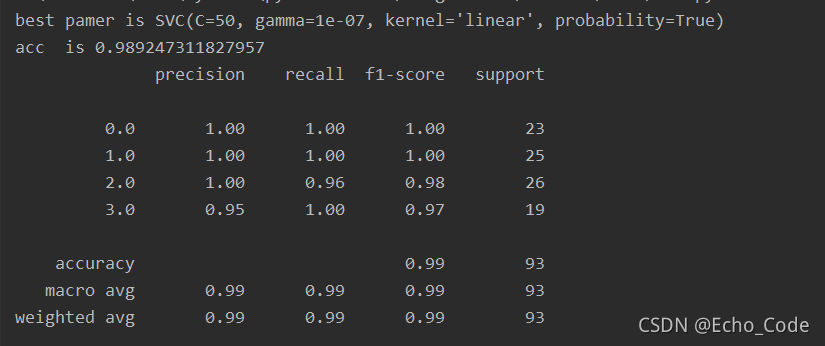
print("best pamer is {}".format(model.best_estimator_))
print("acc is {}".format(acc_test))
print(sm.classification_report(test_y, pred_test_y))
结果如下:
2 .1.2 进行svm的测试
def SVM(train_x ,train_y,test_x,test_y):
clf = svm.SVC(probability=True, C=10, gamma=1e-7,kernel='linear')
clf.fit(train_x, train_y)
pred = clf.predict(test_x)
acc_test = accuracy_score(test_y, pred)
return acc_test
if __name__ == '__main__':
test_ratio = 0.3
tp = 'raw'
X_train, X_test, y_train, y_test = TableDataLoad(tp=tp, test_ratio=test_ratio, start=0, end=404, seed=80)
SVM_Classs_test(train_x=X_train, train_y=y_train, test_x=X_test, test_y=y_test)
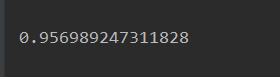
acc = SVM(train_x=X_train, train_y=y_train, test_x=X_test, test_y=y_test)
print(acc)
测试结果